Summary:
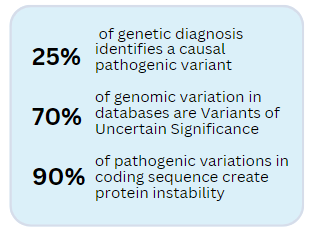
Thanks to the advances in CRISPR technology, the coming decades are poised to usher in the era of personalized medicine. While, on one hand, sequencing techniques have led to huge advancements in understanding rare diseases and their potential treatments, on the other hand, this new technology has also revealed a multitude of Variants with Unknown Significance (VUS), which furthers the challenges involved in a patient’s diagnostic journey. For instance, the Cystic Fibrosis gene (CFTR), first identified in 1989, has 230 variants in CFTR identified as pathogenic, while a much larger 1229 are VUS (Tsui & Dorfman, 2013; Pereira et al., 2019 and ClinVar). In order to develop precision therapeutic interventions for patients in the rare disease space, new tools are needed which can quickly detect pathogenicity and then become cost effective as platforms for discovery of new drugs. The RareResolve Platform we have created for this utilizes gene-humanized animal models as a fast tool for discovery of variant pathogenicity and new therapeutics. In this article, we will examine the journey from unknown disease, to diagnosis, to treatment: often referred to as a patient’s “Diagnostic Odyssey” and “Therapeutic Odyssey”, and discuss how gene humanization using our Whole-gene Humanized Animal Model (WHAM) technology can expedite the drug development process.
Currently, the drug development pipeline is inefficient and costly. The technological advancements of the 21st Century, and the accompanying Genomic Revolution, promised a shift to personalized medicine, where patients would be able to receive targeted therapeutics. Yet technology adoption has taken a bumpy road. Whole Genome sequencing, once rare and expensive, has now become the gold standard and is becoming widespread in its adoption. However, one problem solved is leading to another. The wide spread use of WGS has caused a proliferation of patient data where they often have variants whose pathogenicity is unknown (the “VUS” problem). This outcome is putting even more strain on the drug development industry, where patient inclusion in clinical trials is starting to require that the patient’s variant be assessed as pathogenic (Henrie 2018). The overall result is a pressing need to accelerate drug development process in a patient personalized way – confirm that pathogenicity is caused by the patient variant and then test for drug activity specific to the patient’s variant. Two solutions are evolving to meet this need. One is patient-derived iPSCs to create micro-physiological systems and the other is animal models humanized to contain the patient variant. We take the later approach with our WHAM-humanization technique using the non-traditional animal model of the zebrafish. This achieves the needed high translatability of findings, but at a fraction of the cost of other techniques, and with a speed that can meet the often acute need of gene-encoded diseases.
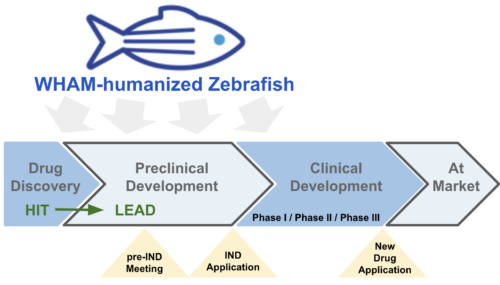
Figure 1. Classic drug development pipeline. WHAM-humanized zebrafish is ideally suited for hit identification, lead maturation, the rapid generation of safety and efficacy data needed for Pre-IND meeting, and diagnostic for setting patient inclusion/exclusion criteria.
For a Patient, It All Begins at the Diagnostic Odyssey:
![]()
For the sick patient, genome sequencing is becoming more common as a tool to aid diagnosis. Often this test identifies a variant in a gene known to be involved in a patient’s disease presentation. Occasionally, these variants are known to be pathogenic. However, more commonly, the variant has never been seen before and is designated as a Variant of Uncertain Significance (VUS). To resolve the uncertainty of these VUS designations, functional tests are needed. When a functional test shows the VUS can cause pathogenic activity, the VUS can be reclassified as a Likely Pathogenic. This designation as a Likely Pathogenic variant allows the patient to become eligible for clinical trials.
For Drug Manufacturers, They Get Involved to Help the Patient Solve the Therapeutic Odyssey:
![]()
For many rare diseases there is no therapeutic option. Yet, because patient populations are often quite small, there is a strong need for affordable approaches to drug development. Direct protein stabilization is a rapidly evolving approach for treating genetic diseases. Many pathogenic variants that occur in a gene lead to protein instability. This protein stability is driven by the composition of its amino acid building blocks. Sometimes changes in these amino acids are well tolerated (benign). However, for an important amino acid, its change to a different amino acid can significantly destabilize the protein and render the protein non-functional (pathogenic). Treatment of a patient with small molecules that stabilize the variant protein can be a successful therapeutic approach. These small molecules bind to the protein and prevent the instability caused by the pathogenic amino acid. As a result, the protein does not degrade and normal function is retained. However, drug development using these approaches needs to be fast and accurate, while at the same time, it needs to be affordable.
Drug Discovery Pipeline
Advances in Artificial Intelligence now allow a protein to be studied at high molecular detail. Clinically-observed variants can be computer simulated for their effect on protein stability. Candidate compounds for protein stabilization can be computer modeled for their ability to reverse this instability. Yet, computer models need to be validated in real-world situations. However, it is unethical to directly test candidates on humans without some prior evidence the candidate compounds can be safe and effective. Rodent and non-human primate models have, until recently, been required for this validation testing (bit.ly/3rbWNEO). However their expense and intrinsic inaccuracies are holding back progress. Often the animal’s sequence for the disease gene (the gene ortholog) has sequence differences in its protein stabilization domains. These amino acid differences can interfere with the desirable drug activity identified in the computer simulations. The result is drugs that might have worked on the human gene will be prevented from working on the the animal’s ortholog version of the disease gene. The candidate compound screened on normal animals, often referred to as “wildtype”, will be labeled as not effective. However this is a false assertion, or a false negative error. On the flip side, compound libraries being screened on wildtype animals will sometimes identify a compound as effective. However, because of human sequence differences, the drugs will not be effective in humans and a false positive error will have been made. Basically, because of genetic sequence differences, drug activity tested in the animal model is often not a good fit to identify the drugs that will work in human.
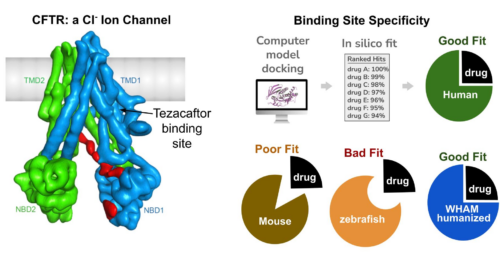
Figure 2. In the CFTR gene of Cystic Fibrosis, the drug component tezacaftor does NOT bind and stabilize the zebrafish version of CFTR gene. Binding Site Specificity: example of how designed molecules will have different binding site capacity between humans and animal models due to differences in primary amino acid sequence. When this amino acid difference is eliminated with WHAM humanization, better quality hits are identified from the animal model.
Pipeline: How Drug Discovery is Improved with RareResolve Platform
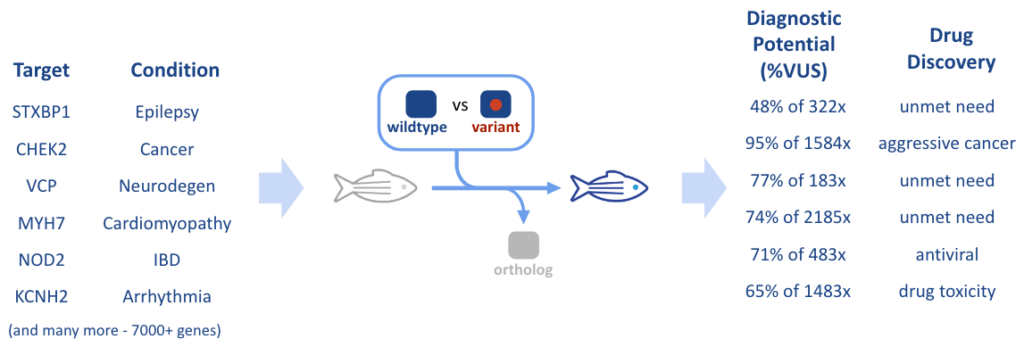
To help speed discovery and provide a more accurate validation of computer-aided drug design, we have developed RarReslove as a more accurate animal modeling platform. We use zebrafish genetically-engineered to express human genes. The method uses Whole-gene Humanized Animal Model, or WHAM, to generate a more accurate model of the patient’s disease state. RareResolve uses the human coding sequence for the target disease gene as a gene substitution for the ortholog – we swap the animal’s version of a gene with the human gene. The RareResolve platform avoids the false positives and false negatives that are inherent in the testing of drug effectiveness in wildtype animals. Drugs that stabilize the WHAM-humanized protein are likely to directly translate to stabilization in the clinic. With the WHAM humanization method, the human gene can be made with and without a pathogenic variant. The resulting set of WHAM-humanized animal models can be used as a tool to diagnose pathogenicity in VUS and to more accurately validate candidate medicines for treatment effectiveness.
Figure 3. RareResolve platform uses Whole-gene Humanized Animal Models (WHAM) for Personalized Medicine Discovery
Real World Example:
Cystic Fibrosis (CF) is caused by pathogenic variants in the CFTR gene and many of these variations cause protein instability. Trikafta is a drug combination that is highly successful in treating this rare disease. Two of the three compounds in the drug are protein stabilizers that prevent protein misfolding and allow the CFTR protein to get to the cell surface and properly function as an ion channel. In selection of the compounds for stabilizing the protein, the binding site for protein stabilization was found to occur at an inherently unstable position in the protein. Small molecules can bind this site and stabilize the protein. Premature protein degradation is avoided with stabilizer treatment and the CFTR protein can properly traffic to its standard location and perform its normal function in regulating lung mucus formation. The result with drug treatments is appropriate levels of mucus form on lung tissue and tissue inflammation is prevented. Trikafta has been a game changer for the CF community and now kids have access to a drug that allows them to proceed with a normal, physically-active life.
Conclusion:
The RareResolve platform is well suited to the evolving needs of the rare disease community. By inserting a human gene as a gene replacement into the zebrafish genome, the resulting WHAM-humanized platform generates higher accuracy in drug discovery efforts. A computationally-identified hit can be tested in a system with greatly reduced risk of false positive and false negative outcomes. Further, the unique attributes of a zebrafish, such as larval screening in 96-well plates, make the animal well suited to the medium throughput needed of screening 100s of AI-identified hits. Using WHAM humanized animals, we will be looking for drug activity as a chemical rescue experiment – we will be identifying which leads are capable of reversing the disease activity of a pathogenic variant. Intriguingly, groups of pathogenic variant animals can be prescreened to identify the patient variants that are the responders. Analysis of multiple animals as Clinical Avatars of the patient condition will greatly improve clinical trials. Patient inclusion/exclusion criteria can be set by observing which variants are responsive to a candidate drug. As a result, rapid animal model creation with the RareResolve platform is starting to provide a fast, accurate, and affordable system for today’s personalized medicine needs.
‘Contact our experts to discuss how we can accelerate your rare disease therapeutic discovery
References:
- Tsui, L. C., & Dorfman, R. (2013). The cystic fibrosis gene: a molecular genetic perspective. Cold Spring Harbor perspectives in medicine, 3(2), a009472. https://doi.org/10.1101/cshperspect.a009472
- Pereira, S.VN., Ribeiro, J.D., Ribeiro, A.F. et al. Novel, rare and common pathogenic variants in the CFTR gene screened by high-throughput sequencing technology and predicted by in silico tools. Sci Rep 9, 6234 (2019). https://doi.org/10.1038/s41598-019-42404-6
- Henrie A. et al. ClinVar Miner: Demonstrating utility of a Web-based tool for viewing and filtering ClinVar data. Hum Mutat. 2018 May 23. doi: 10.1002/humu.23555. https://www.ncbi.nlm.nih.gov/pubmed/29790234