CRISPR Knock-in Services: Customized Gene Editing Solutions for Precise Modifications
Experience the groundbreaking potential of CRISPR-Cas9 technology, revolutionizing the world of genetic engineering. At InVivo Biosystems, we specialize in providing cutting-edge solutions for precise DNA sequence editing in a wide range of organisms, including the development of custom CRISPR knock-in models through gene knock-in cell lines technology.
Our expertise extends to the highly versatile nematode Caenorhabditis elegans, a key player in genetic and biomedical research. From point mutations to floxed alleles, degron tagging to fluorescent and immunotagging, we have you covered, employing CRISPR knock-in instruction and CRISPR knock-in techniques for tailored genetic alterations.
More C. elegans Models Services
Applications of CRISPR Knockin
- Modeling genetic diseases in mammalian cells and human cells.
- Studying the effects of disease-causing mutations on gene expression and function.
- Developing transgenic animals that express a specific gene or protein, leveraging CRISPR knock-in cell lines technology.
- Creating animal models that accurately mimic human disease states.
- Performing site-specific modifications in the genome, such as introducing reporter genes or modifying regulatory sequences.
- Studying gene regulation and expression with greater precision and accuracy
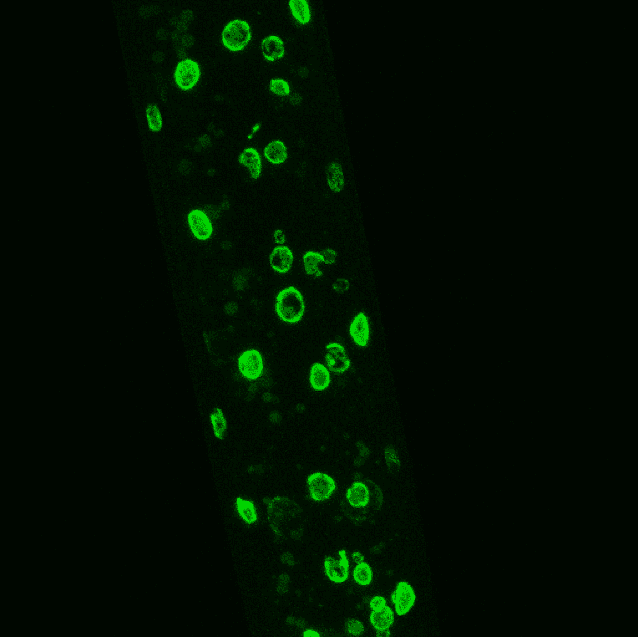
Image courtesy of Dr. André Catic, Baylor College of Medicine.
Fluorescent Tagging of Intestinal nuclei from late L4 worms in an lmn-1p::GFP::LMN-1 transgenic strain. The image is lmn-1 tagged endogenously with GFP.
Applications of CRISPR Knockin
CRISPR knockin technology is a valuable tool for various biomedical research applications, including the modeling of genetic diseases in mammalian cells and human cells. By introducing disease-causing mutations into a specific genomic locus with high knock-in cell lines efficiencies using embryonic stem cells, researchers can study the target effects of the mutation on gene expression and function.
In addition to disease modeling, CRISPR knockin can be used to develop transgenic animals that express a specific gene or protein. This method utilizes the precise targeting of the target locus, allowing for the creation of animal models that accurately mimic human disease states.
Furthermore, CRISPR knockin can be utilized for site-specific modifications in the genome, such as introducing reporter genes or modifying regulatory sequences. This enables researchers to study gene regulation and expression with greater precision and accuracy.
At InVivo Biosystems, we specialize in providing high-quality CRISPR knockin services for a range of research applications. Our experienced team utilizes cutting-edge technology to achieve high knock-in efficiencies and precise targeting of the target locus, ensuring accurate and reliable results.
Types of C. elegans Knock-in Services
Our C. elegans CRISPR knock-in services provide researchers with tailored solutions for precise genetic modifications in C. elegans.
Fluorescent Tag

Visualize your protein via the addition of a fluorescent protein tag at the native locus.
Immunotag

Use your protein for biochemical studies by adding an immunotag at the native locus.
Point Mutation

The best gene editing method for creating small, precise edits to introduce a small number of nucleotide changes at a target site.
Whole Gene Humanization

Replace a C. elegans gene with an orthologous gene from human or any other organism.
C. elegans Knock-in Service Custom Offerings
Our C. elegans Knock-in service custom offerings provide researchers with tailored solutions for precise genetic modifications in C. elegans.
Build Type
Full Build
Candidate Lines
Custom Injection Mix
Transgenic Design
Injections
Candidate Screening
Strain Confirmation
Pricing
$3,465 and up
$2,435 and up
$1,740 and up
Timeline
8 weeks and up
6 weeks and up
4 weeks and up
Note: Additional charge applies for cargo size above 200bp for all packages.
C. elegans Knock-in Service Details
Here are the key details of our C. elegans Knock-in Services:
C. elegans Point Mutation Service
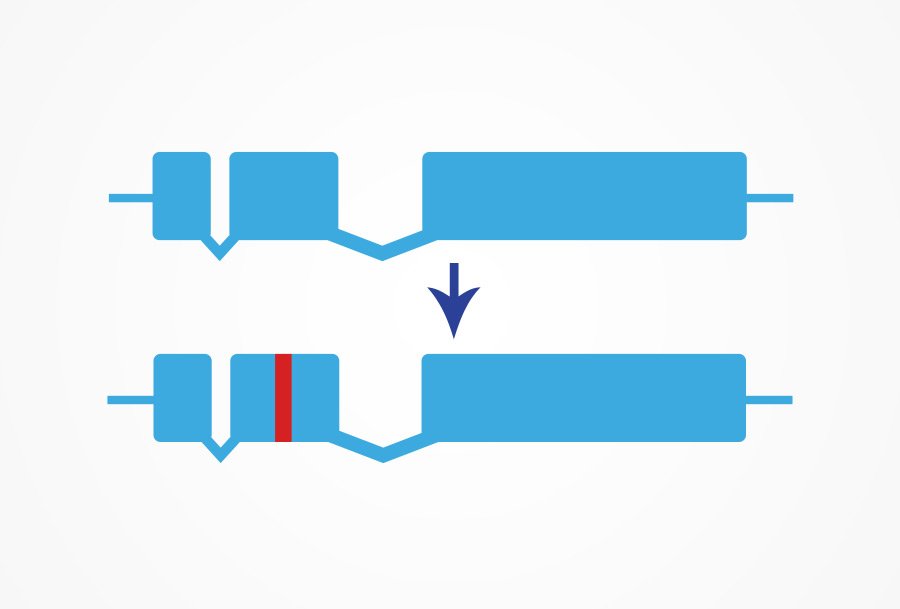
Our Point Mutation service utilizes the cutting-edge CRISPR/Cas9 gene editing services method to introduce precise, small edits to a target site. It enables us to achieve high levels of specificity and accuracy, resulting in reliable and reproducible results. With our Point Mutation service, researchers can accelerate their research and gain a deeper understanding of the function and behavior of genes. We provide comprehensive end-to-end support, from project design and consultation to final delivery of the desired point mutation.
- Study a disease-causing mutation
- Humanize critical amino acids
- Explore enzyme binding sites
- Introduce phosphomimetics
- Mutate isoform start sites
- Make any specific mutation of interest
CRISPR/Cas9 technology enables us to achieve high levels of specificity and accuracy, resulting in reliable and reproducible results. With our Point Mutation service, researchers can accelerate their research and gain a deeper understanding of the function and behavior of genes. We provide comprehensive end-to-end support, from project design and consultation to final delivery of the desired point mutation.
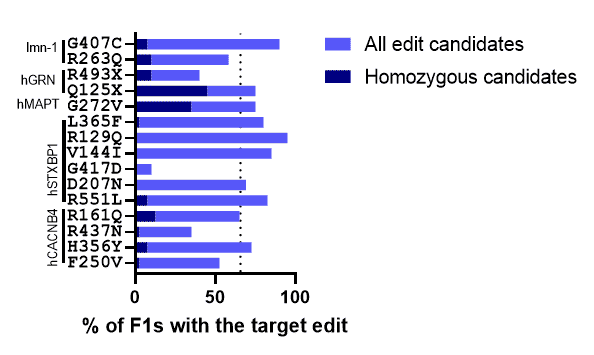
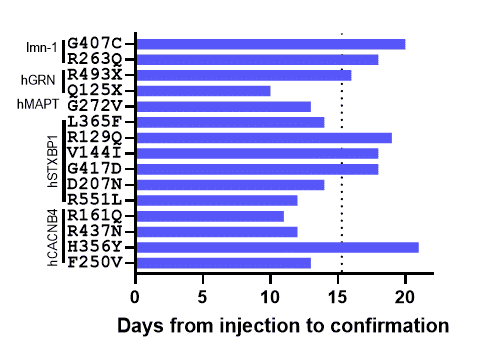
96 Point Mutations In 1 Gene
All 96 mutations were created in the STXBP1 gene (which is associated with epilepsy in humans) via CRISPR. The worm homolog of STXBP1, unc-18, causes uncoordination and near-complete lack of pharyngeal pumping when knocked out. The functionality is restored by replacing the worm gene with the coding sequence for human STXBP1.
C. elegans Fluorescent Tagging Service
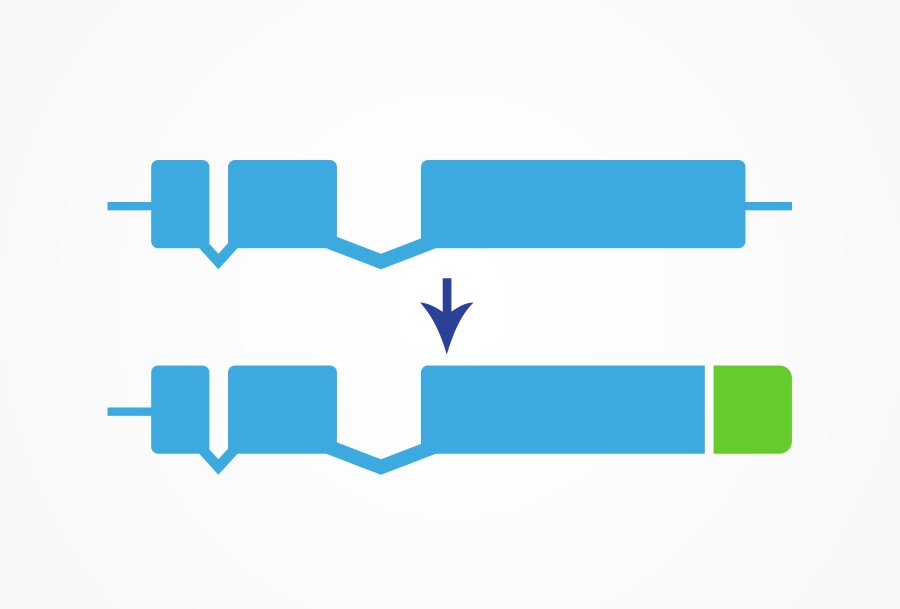
Fluorescent tagging of genes is a widely used technique in many model organisms to study gene expression patterns and protein localization. In C. elegans, this technique is particularly effective due to the worm’s transparency. Our Knock-in Fluorescent Protein service uses CRISPR gene editing technology to add a fluorescent protein tag at the endogenous locus.
- Detect protein expression levels in-vivo at native gene expression levels
- Ensure that you have the correct expression pattern
- Identify protein-protein interactions
- Confirm protein degradation when paired with a degron sequence
- Visualize protein synthesis rates
- Quantify protein levels
- Visualize protein localization to organelles or membranes
You can choose from a variety of optimized fluorophores, including YFP, BFP, eGFP, mCherry, mOrange, mScarlet, and more. Our service also offers the Fluorescent Protein RNAi technique to reduce or eliminate fluorophore expression.
Concerned about the impact on protein function? We can use a 2A or SL2 sequence to separate the components. Our team provides end-to-end support, from project design and consultation to final delivery of the desired fluorescently tagged strain.
Here are the things to consider when ordering your strain:
- Fluorescent protein best suited for your experiments
- Best terminus to tag
We are not limited to a fixed list of fluorophores. If you have a fluor that you would like to use, you can provide us the sequence and we will incorporate it into the design of your transgenic project. If you need your fluorescent transgenics lines to be imaged, we can readily help you with that too! If you are unsure, our genetic engineering experts will be glad to advise you.
C. elegans Immunotag Service
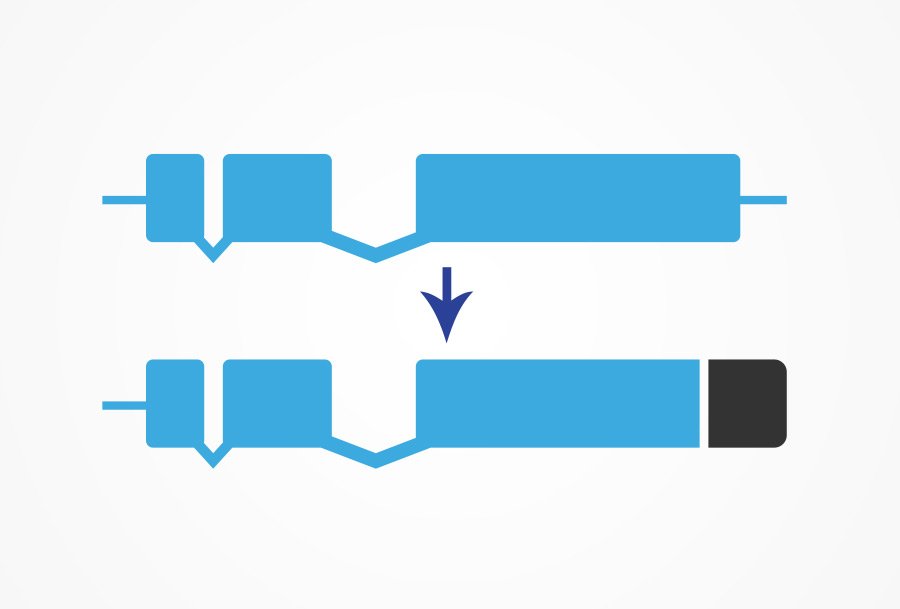
The inclusion of an immunotag at the endogenous locus provides the opportunity to accurately quantify protein levels without causing any interference in the gene expression levels.
Our Immunotagging service utilizes the highly advanced CRISPR/Cas9 gene editing technology to add immunotags to your protein of interest. You may select from a wide range of tags, such as FLAG, HA, HIS, TAP, or S-peptide, to suit your requirements.
All of these tags have undergone rigorous optimization to ensure their successful expression in C. elegans.
C. elegans Degron Tagging Service
Our degron tagging service offers the option to knock-in a degron tag on an endogenous protein or fluorescent protein. Adding a degron tag provides several benefits, such as using protein degradation as a complementary method to genetic knockouts or RNAi, tagging your protein of interest for degradation at the protein level, controlling the timing of protein degradation with an inducer (e.g. auxin, blue light), and confirming protein degradation when paired with a fluorescent sequence.
Our degron tags can be used in combination with fluorescent tagging to visualize protein expression and degradation.

Our service allows you to select from a range of degron inducible and developmental systems. These include Auxin Inducible Degradation, Photosensitive or blue-light inducible degradation, degron-tagged reporters of membrane topology, and OMA-1 tags that work to degrade protein from the 1-cell stage.
Things to consider when ordering your strain:
To ensure optimal results, consider the following factors when placing your order:
- Degron tag best suited for your experiments
- Best terminus to tag
- For auxin-inducible degradation, TIR1 is necessary to generate a functional recognition complex and achieve successful target degradation. If you do not have a TIR1-expressing strain ready, we can suggest the most suitable commercially available strain, or we can generate a custom TIR1 strain for you.
Our degron tags can be used in combination with fluorescent tagging to visualize protein expression and degradation. If you have any further queries, please do not hesitate to contact our team for assistance.
C. elegans Whole Gene Humanization
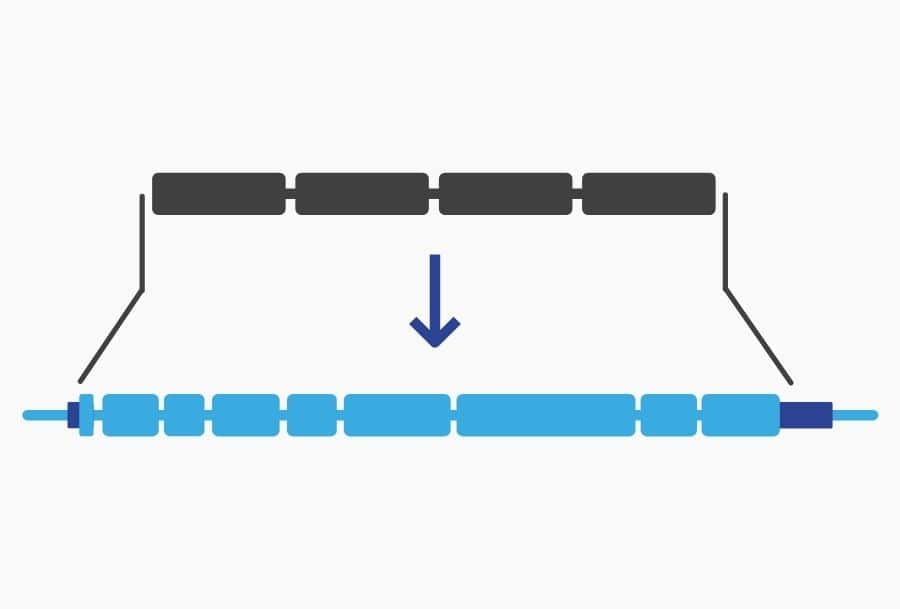
Whole gene humanization allows you to replace a C. elegans gene with an orthologous gene from human or any other organism. If the human gene rescues the function of the gene deleted-KO, you know there is conserved biology. This is often the first step in a project to create a model to study clinical variants.
We take care to design our humanization projects to preserve endogenous transcription signals so the expression patterns and levels will be maintained. In addition, we have a proprietary sequence optimization protocol that we use to create a transcript that will be expressed well in C. elegans.
Creating a whole gene humanized line enables you to:
- Determine if the human protein is functionally equivalent to the C. elegans protein.
- Study drug effects on the human protein in an animal model.
- Create a model for the study of clinical variants.
C. elegans Whole Gene Humanization Service Pricing
Service Package
Price
Est. Delivery Time
Full Build
$5,105 and up
8 - 12 Weeks
Candidate Lines
$4,024 and up
6 - 8 Weeks
Custom Injection Mix
$2,295 and up
4 - 6 Weeks
Note: Additional charge applies for proteins over 2000 amino acids in size for all packages.
Choose From a Variety of Fluorophores
You can choose from a variety of fluorophores including YFP, BFP, eGFP, mCherry, mOrange, mScarlet, and many more. All of our fluorescent fluorophores are optimized for expression in C. elegans via codon optimization and insertion of introns.
Use our fluorescent fluorophore tags in combination with the Fluorescent Protein RNAi to reduce or eliminate fluorophore expression.
Concerned that the addition of a fluorescent fluorophores will inhibit protein function? We can use a 2A or SL2 sequence to separate the 2 components!
Other Gene Editing Service We Provide
CRISPR Knockin Services FAQs
Here are some frequently asked questions about CRISPR knockin.
CRISPR knockin involves introducing a specific DNA sequence into a targeted genomic locus, while CRISPR knockout involves disrupting or deleting a specific gene or genomic locus.
CRISPR knockin can be used to model genetic diseases, develop transgenic animals, and create site-specific modifications in the genome for studying gene regulation and expression.
The design of a CRISPR knockin experiment depends on the specific application and desired outcome. Consult with a CRISPR Expert to guide your experiment.
Successful CRISPR knockin can be confirmed using various methods, including PCR, sequencing, and fluorescent assays.
Yes, CRISPR knockin has potential for therapeutic applications, such as introducing therapeutic genes or correcting disease-causing mutations in patient cells.
Ready to get started?
Ready to connect with us to learn more about working with our company or our technology?
Submit your inquiry below & we will get back to you soon.