Summary
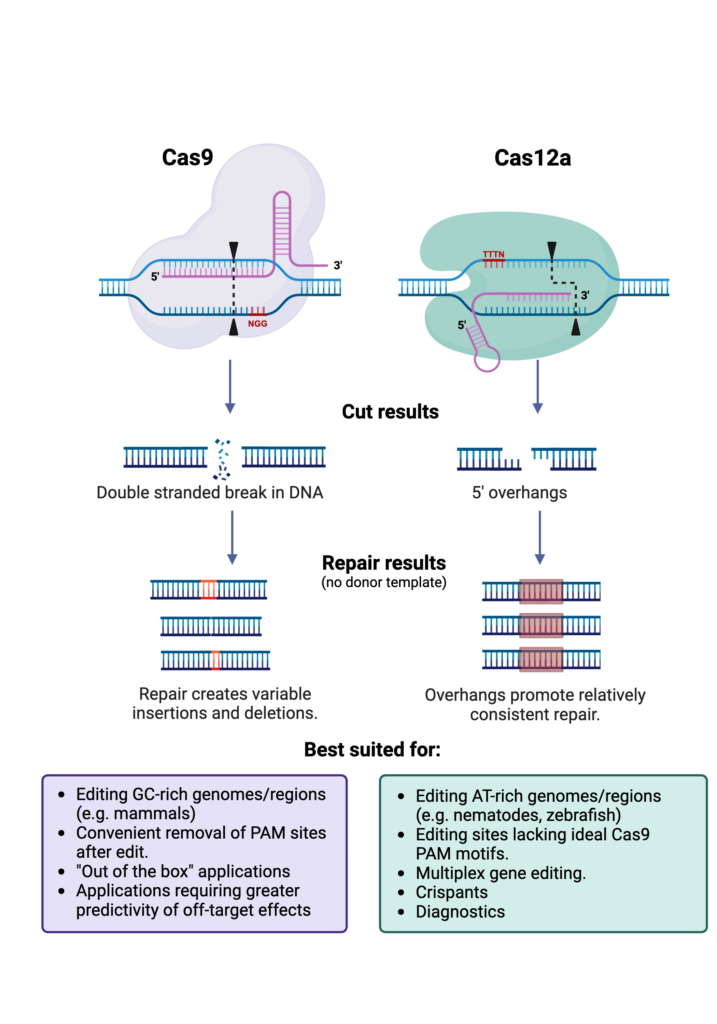
- Cas12a’s sequence recognition motifs allow access to AT-rich genomes and loci that are less amenable to Cas9 gene editing, showcasing efficient genome editing capabilities.
- Cas12a cutting of DNA leaves overhangs that produce more uniform lesions than Cas9 and might be used to favor homology-directed repair (HDR), a process crucial in minimizing target effects and enhancing genome editing applications.
- Cas12a nuclease can facilitate multiplex gene editing or Crispant capabilities, significantly advancing the field of mammalian cells and plant genome editing. Pushing forward the envelope in agricultural advancement through precise genome editing applications.
Introduction
The story of CRISPR-Cas9 exemplifies how delving into the richness of biodiversity can uncover transformative tools in unexpected places, including the exploration of the prokaryotic genome and the bacterial genome. With the rapid ascension from that first Cas9 system engineered for genome editing in 2012 to the Nobel Prize in 2020 to an incoming wave of novel therapies and diagnostics, we might expect that the CRISPR story is one where only the first chapter has been written. When looking back on some of our more challenging gene edits, we asked whether new technologies, such as Cas12a might have made those edits easier, especially in the context of editing efficiency and addressing potential targets.
With a shorter history and fewer publications, some researchers might be hesitant to incorporate Cas12a into their approaches, despite its potential for enhanced genome editing specificity and significant implications for genome editing abilities in both human cells and plant genomes. We previously took a detailed look at the development history, mechanistic differences, and advanced applications in a previous post titled “History of CRISPR: Cas9 and Cas12”. Here, we share some of the basics of what Cas12a has since taught us about this new chapter in genome editing.
Class of Cas
To understand the significance of Cas12a, it helps to understand that CRISPR-Cas systems have deep evolutionary roots and diverse functionalities, serving as an ancient form of adaptive immunity in microbial defense. The most well-studied CRISPR-Cas systems so far are an ancient form of the adaptive immune system used by bacteria and archaea to fend off invading viruses. Canonically, a CRISPR-Cas system comprises a genomic region of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and one or more CRISPR-associated (Cas) proteins that edit and interpret viral signatures stored within the repeats. The extensive repository of microbial genomes, coupled with cutting-edge predictive tools, enabled scientists to uncover a diverse array of CRISPR-Cas systems. Among these systems, a specific “Cas9” protein found its place in the spotlight – a protein derived from Streptococcus pyogenes with a desirable nuclease activity. This protein became the linchpin for the gene editing revolution, laying the groundwork for the transformative power of CRISPR. The impact of Cas9 on gene target editing and therapeutics spurred researchers to dive deeper into the microbial treasure trove, leading to the discovery of Cas12a in 2015.
Enter Cas12a
Discovered in 2015, Cas12a marked a new chapter in genome editing. The protein structures of Cas9 and Cas12a, though sharing functional similarities, suggested a long history of independent evolution. A key innovation that enabled Cas9 to edit any DNA target sequence was the joining of two RNA elements, the CRISPR-RNA (crRNA) and theTrans-activating crRNA (tracrRNA) into a single molecule. One feature that sets Cas12a apart is its more minimalistic system, as it processes its own crRNA and does not require a tracrRNA sequence. This reduces the RNA payload that must be delivered to cells along with the Cas protein. Both enzymes require a protospacer adjacent motif (PAM) sequence for binding to target DNA, but the PAM sequences are different in both the position and the base composition, imparting each with complementary access to different types and regions of genomes. The cutting mechanism differs as well. Standard Cas 9 cleaves straight through double-stranded DNA leaving blunt ends, whereas Cas12 leaves overhangs that help guide more predictable repair processes. How do these properties impact genome editing abilities and how do you select your strategy? Let’s discuss some of the key points.
Alternate Recognition Sequences Unlock Access to More Genes
We were introduced to Cas12a by asking whether new CRISPR-based tools might help improve our editing of difficult loci with a dearth of optimal Cas9 cutting sites. This inspired us to retrospectively examine whether designing with Cas12a instead of Cas9 would have improved our results on some of these more challenging types of edits. With bioinformatic tools that have expanded to incorporate Cas12a motifs, we scanned the locus and found predicted Cas12a guide RNA recognition sequences in ideal positions. Whereas Cas9 recognizes 5′-NGG-3′ PAM sequences, Cas12a recognizes AT-rich sequences with a PAM typically 5′-TTTV-3′, expanding the horizon of genome editing to AT-rich genomes and loci that might be intractable with Cas9. This capability holds particular significance in applications such as genome editing in zebrafish or nematodes, where the prevalence of AT-rich sequences frequently poses a challenge. To the benefit of anyone designing these guidest, the bioinformatic tools for designing sgRNAs and Cas12a edits have been well developed, with the caveat that prediction of off-target activity still lags behind Cas9. Furthermore, the enzyme and reagents are commercially available from common suppliers. With a few minor adjustments to our workflow, anyone could use this new approach and be as time efficient and accurate as standard Cas9 edits.
Distinct Cutting Mechanism Opens up New Applications
Our initial Cas12a explorations prompted us to ask what other possibilities might be unlocked by the use of Cas12a. There are several mechanistic differences that open up possibilities for Cas12a in gene editing as well as gene therapy, and diagnostics [link to previous post], but one of the more prominent features is Cas12a’s staggered cutting mechanism. Although different cutting functions have been engineered into versions of Cas9, typically Cas9 will introduce a single double-stranded break at a cut site that is then repaired by non-homologous end joining (NHEJ) using the cell’s own machinery. However, the cut placement and repair process have some variability that creates a heterogeneous pool of edits. In contrast, Cas12a produces a staggered cut with a 5′ overhang that can be used to guide the repair process. Whereas the blunt ends of a Cas9 cut are frequently chewed back or frayed in the repair process, the sticky ends produced by a Cas12a cut tend to guide a more consistent and predictable repair. When examining repaired cut sites, almost no two Cas9 isolates will have the same target sequence at the repair site, unless there is a local microhomology templating the repair. On the other hand, for cuts made with Cas12a, the repaired sequences tend to be uniform across most isolates. For standard edits involving homology-directed repair, the variability of Cas9 cuts is inconsequential due to the need to ultimately screen for desirable edits. However, for some novel extensions of CRISPR technology discussed below, the cuts need to be consistent enough to obviate the need for intermediate screening.
Multiplexing and Crispants
As single-gene edits using CRISPR-Cas9 became commonplace, it was not long before the need arose for multi-loci or multi-gene edits. The heterogeneous repair of Cas9 cutting sites as well as the need to introduce large guide RNAs makes Cas9 less than ideal for editing more than one gene simultaneously. With a more uniform cutting mechanism, Cas12a stands as a stronger candidate to lead the way with multiplex gene editing. For emerging applications, such as gene therapy, it may be advantageous to be able to edit multiple loci simultaneously without having to individually confirm each edit. In being a slightly smaller protein and requiring a smaller cr-RNA that lacks a tracrRNA, Cas12a further facilitates multiplex gene editing by reducing the overall payload, possibly promoting more consistent delivery to targets.
The power to multiplex gene edits is also transformative for the application of Crispants, a type of rapid gene knockout using CRISPR-Cas technology [link to Crispant page]. Whereas a typical CRISPR edit requires months of propagation and screening of injected animals to confirm the edit, a Crispant approach delivers a highly penetrant gene knockout in the injected or F0 generation to enable rapid target screening and validation. The key to generating a complete gene knockout so early lies in simultaneous targeting of multiple loci within a gene. The Crispant approach can be advantageous for rapid early target validation and viability testing in preclinical therapeutic studies.
Benefits of an Expanded Toolkit
The introduction of Cas12a into the CRISPR toolbox extends the range of possibilities for researchers. While Cas12a and Cas9 share functional similarities, their differences are profound and significantly impact their application’s diverse range of editing efficiencies, target sequences, and genomic contexts. The distinctions encompass mechanisms, RNA processing, PAM recognition, target DNA binding, and catalysis. The distinct PAM recognition sequences of Cas9 and Cas12 influence their applicability, offering researchers greater flexibility in designing experiments tailored to their specific needs. As a skilled cabinet maker will choose appropriate joints to assemble the most durable drawers, a biologist benefits from having a variety of molecular joints for assembling DNA. Cas12a’s ability to target AT-rich sequences opens up new frontiers in genome editing, enabling precise modifications in previously challenging genomic landscapes. Its self-recycling endonuclease activity may contribute to decreased non-homologous end joining (NHEJ) and increased homology-directed repair (HDR) events, offering a refined approach to genetic edits.
The Future of Cas12a
Given the longer history and continued advancements using Cas9 gene editing, we expect that there will be much more to see from Cas12a in the near future. These adventures with Cas12a have begun to give us an understanding of the nuances that influence the choice between Cas9 and Cas12a. Although the tools are not yet as elaborate as with Cas9, there are ample bioinformatic resources and reagents currently available that should encourage any researcher to experiment with Cas12a.
Is Cas12a Right for your Research?
Reach out to InVivo Biosystems today to see how we can help you meet your research needs.
References:
- Ebbing, A., Shang, P., Geijsen, N. & Korswagen, H. Extending the CRISPR toolbox for C. elegans: Cpf1 as an alternative gene editing system for AT-rich sequences. MicroPublication Biol. (2017) doi:10.17912/W2237D.
- Makarova, K. S. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020).
- Paul, B. & Montoya, G. CRISPR-Cas12a: Functional overview and applications. Biomed. J. 43, 8–17 (2020).
- Zetsche, B. et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell 163, 759–771 (2015).
- Zetsche, B. et al. Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34 (2017).


