Between the early 2000s and mid-2010s, artificial enzymes engineered to manipulate genomes – Zinc-finger nucleases (ZFNs) and transcription-activator–like effector nucleases (TALENs) – had garnered considerable interest in the genome editing community. Starting in 2012, the CRISPR-Cas system was developed into a revolutionary molecular tool for DNA and RNA “surgery”. Clustered regularly interspaced short palindromic repeats (CRISPRs) are a family of nucleotide sequences present in multiple copies in the genome. Those sequences are called DNA direct repeats. Thanks to these patterns, CRISPRs can be used as a way to find a specific sequence of DNA inside a cell, or to shut one off by targeting and cutting a specific gene.
In the last decade, scientists have focused on the Cas endonucleases in an attempt to continuously improve their efficiency as genome editing tools. By better understanding the mechanisms of Cas proteins, scientists are able to leverage them for various applications. Despite their short history, CRISPR-Cas systems and Cas proteins have already been the subject of over thirty thousand publications1.
In this blog post, we summarize the history of two of the most widely studied Cas proteins and how they are shaping the future of research and medicine.
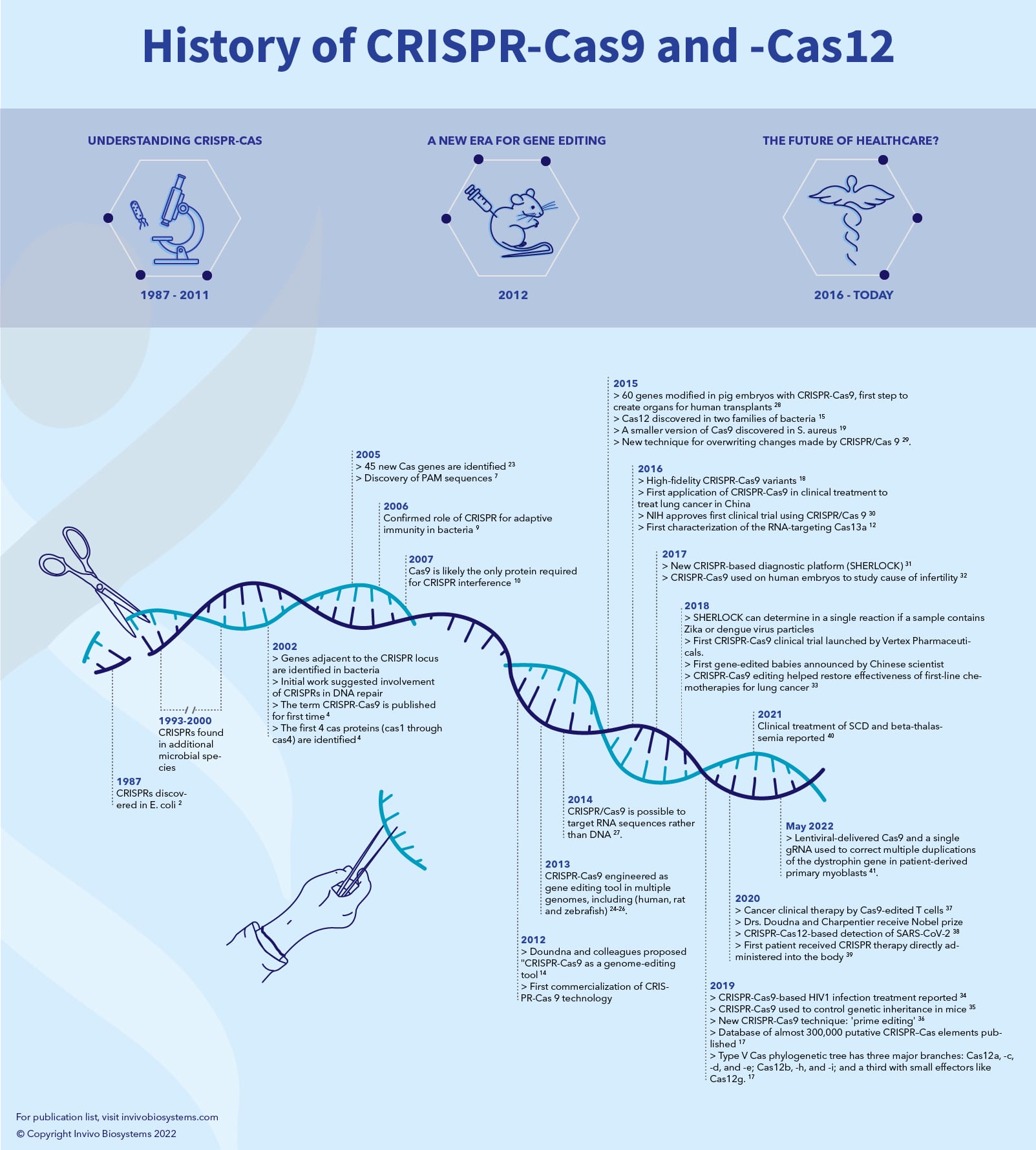
Discovery of CRISPR
In 1987, while studying a gene responsible for the conversion of alkaline phosphatase, Dr. Yoshizumi Ishino and his team from the University of Osaka observed an “unusual arrangement with repeated sequences” in the bacteria E. coli2. However, due to the lack of sufficient DNA sequence technology and data (the sequencing process had not yet been automated), the function of these arrays remained a mystery. It wasn’t until 1993 that Dr. Francisco Mojica, then a doctoral student at the University of Alicante in Spain, characterized for the first time what is now called a CRISPR locus3. Almost a decade later, in the year 2000, Mojica reported that the different repeat sequences that he had observed shared a common set of features. In collaboration with Ruud Jansen from the University of Utrecht in the Netherlands, the term CRISPR was invented. Jansen was the first to publish these findings in 2002, as he identified the first four cas (CRISPR-associated) proteins (cas1 through cas4)4. He notes that those cas are present in prokaryotes (Archaea and Bacteria), but absent from eukaryotes or viruses. In 2005, Mojica reported that these sequences matched snippets from the genome of bacteriophage5. This finding led to the hypothesis that CRISPR is an adaptive immune system5-7.
What is The CRISPR-Cas System?
The role of CRISPR systems in adaptive immunity was pursued by various teams in the late 2000s8,9 until Philippe Orvath and colleagues finally demonstrated it empirically in 200710. They showed that cas genes do not directly provide resistance but rather mitigate the insertion of additional fragments of foreign DNA (spacers), as well as repeats. The CRISPR spacers are used as immunological recordings to combat future infections.
The Cas protein sequences and the genomic organization of CRISPR-cas loci display a wide diversity and are rapidly evolving. Since the discovery of CRISPR, 2 classes, 6 major types and more than 33 subtypes of Cas proteins have been identified11. Thanks to the screening of constantly updated genomic and metagenomic databases, our knowledge of this diversity of Cas proteins is ever expanding. For example, we know that while some Cas proteins like Cas9 cleave DNA, others, like Cas13 cleave RNA12,13.
Cas9: The Original CRISPR Enzyme
The best known Cas protein to date is SpCas9, the original gene editing enzyme. Cas9 is found in several strains of bacteria, where it originally evolved to cut invading foreign DNA. In 2005, Alexander Bolotin revealed an unusual CRISPR locus while studying the newly sequenced bacteria Streptococcus thermophilus7. Although the CRISPR array (alternating conserved repeats and spacers) was similar to previously reported systems, it lacked some of the known cas genes and instead contained novel cas genes.
One of these new cas genes encoded a large protein predicted to have nuclease activity, Cas9. Bolotin also noted that the spacers, which had homology to viral genes, all shared a common sequence at one end. This piece of targeting DNA located next to the desired cleavage site is called the protospacer adjacent motif (PAM) and is required for target recognition. Bolotin’s work on bacterial immunity ultimately led to the groundbreaking discovery by Jennifer Doudna and her team that Cas9 is indeed an RNA-guided endonuclease14. Based on this discovery, Doundna and colleagues proposed “an alternative methodology based on RNA-programmed Cas9 that could offer considerable potential for gene-targeting and genome-editing applications”14.
Cas12: The Future of CRISPR-Dx?
Discovered in bacteria in 201515, Cas12 is a more recent addition to the CRISPR family. Yet, in the last couple of years, Cas12 has gathered growing interest for its potential use in diagnostics16. Contrary to Cas9, which requires two RNAs to recognize and cut its target, and possesses no inherent RNAse activity, Cas12 processes its own guide RNAs (single RNA-guided endonuclease) and only requires crRNA for targeting.
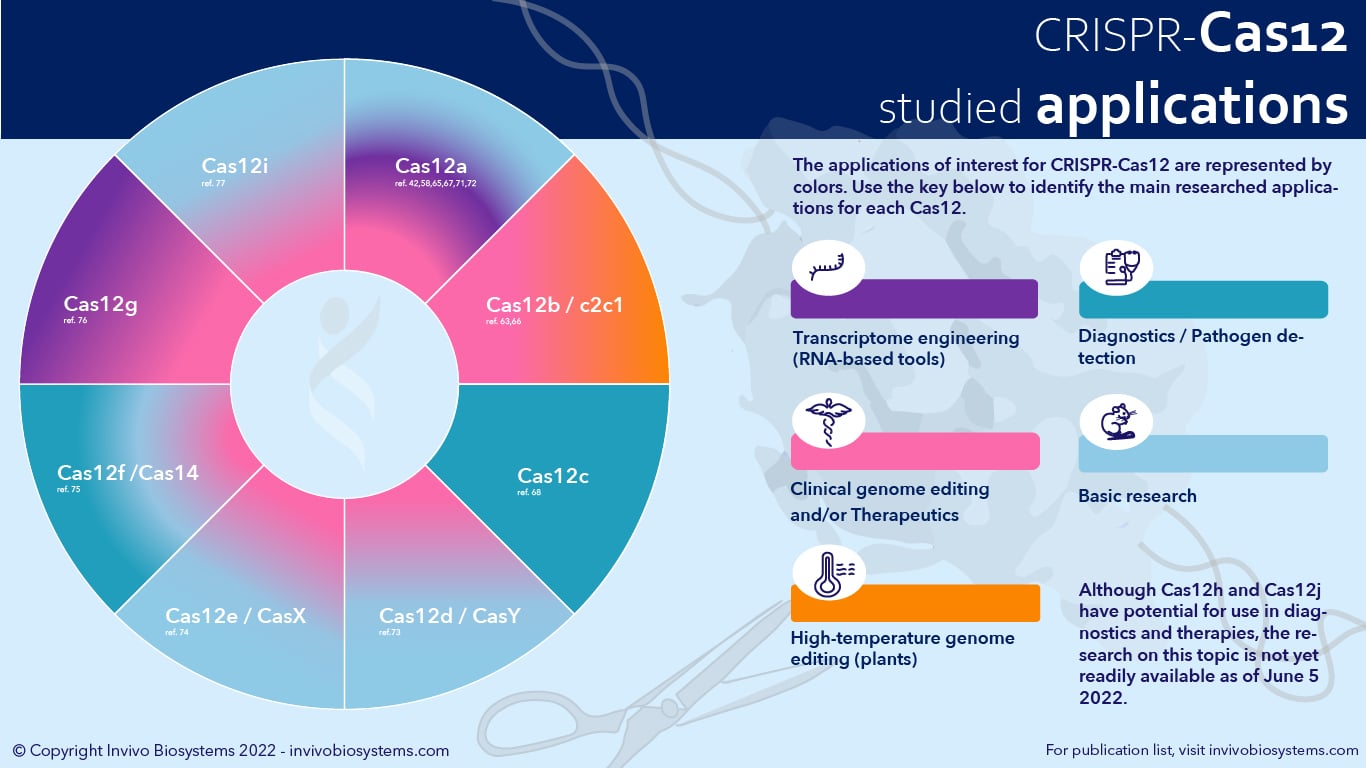
David Scott and colleagues from Arbor Biotechnologies, in collaboration with Eugene Koonin, recently studied the entire Cas12 family to find effectors with new functionality17. Together, they built and searched a database of almost 300,000 putative CRISPR–Cas elements for type V systems. They then discovered that the type V phylogenetic tree has three major branches: one with Cas12a, c, d, and e; a second that includes Cas12b, h, and i; and a third with smaller effectors such as Cas12g. Interestingly, Cas12g can cleave both single-stranded DNA and RNA in trans in a non-specific manner. This ability could be exploited to detect viruses or the presence of certain mutations. Cas12i, on the other hand, does not need a trans-activating CRISPR RNA (tracrRNA) for single-strand DNA cleavage and is able to introduce a break only in the one strand of DNA that is not paired with the guide RNA. Because of this, Cas12i has the potential to become a highly specific platform for therapeutic genome editing. Despite their promising capabilities for diagnostics and therapeutics (see infographic below), the short history of Cas12 endonucleases often makes them less attractive to researchers looking for efficient and precise genome editing tools.
Table 1: Comparison of Cas9 and Cas12a
| Cas9 (also known as SpCas9) |
Cas12 (also known as Cas12a or Cpf1) |
Notes | |
| Year discovered | 2012 | 2015 | Cas 9 is the go-to option for genome editing thanks to its longer history and more publications. |
| Number of publications (as of May 2022) | 20,100 | 700 | |
| Patent rights | Broad Institute | Broad Institute, MIT, and Harvard | |
| Cas family type | Type II | Type V | |
| Size (amino acids) | 1,000-1,600 | 1,300 | Cas12’s smaller size allows it to be more easily delivered inside cells using common viral vectors like adeno-associated viruses (AAVs). |
| Nuclease activity | Single | Dual | The dual nuclease activity of Cas12a is essential for its ability to create double strand breaks in the DNA and is dependent on the RuvC domain. |
| RNA(s) needed | crRNA + tracrRNA (or single-guide RNA) | crRNA | Cas12a processes its own pre-crRNA into mature crRNAs, without the requirement of a tracrRNA, making it a unique effector protein with both endoribonuclease and endonuclease activities. This dual nuclease activity is advantageous for multiplex gene editing, transcription, epigenetic modulations and base editing. |
| Nuclease site | 2 nuclease domains: HNH and RuvC | Single nuclease site RuvC-Nuc | |
| Non-specific ssDNA cleavage | No | Yes | Once the complex Cas12/crRNA/target DNA is established, the non-specific cleavage of any collateral single-stranded DNA (ssDNA) is induced. This activity can be exploited to create biosensing systems that merge Cas12 effectors with amplification methods to enable rapid and specific detection of pathogen DNA samples. |
| Type of cut in dsDNA | Blunt cut | Staggered 5′ overhang | This feature facilitates site-directed integration, making Cas12a a useful tool similar to restriction enzymes for precise in vitro DNA assembly. Cas9’s blunt double-stranded may also induce more aberrations during repair. |
| Trans cleavage | No | Yes | The non-specific single-stranded DNA (ssDNA) cleavage activity in Cas12a is sufficient to completely degrade both linear and circular ssDNA molecules. |
| Seed sequence | 10 nt | 5-6 nt | |
| PAM sequence recognized | 3′ G-rich 3-5 nt |
5′ T-rich | Most mammalian genes are GC-rich, so finding the requisite GG in mammalian cells for targeting Cas9 to a specific location is easier than for Cas12. Cas9’s preference for GC-rich PAMs limits the targeting of AT-rich sequences, for example most of the non-coding genome in zebrafish is AT-rich. |
| Specificity in vivo | + | ++ | |
| Mismatch tolerance in vivo | + | ++ | CRISPR nuclease tolerates some levels of imperfect complementarity between gRNA spacer sequences and protospacer sequences of the targeted genome. A recent comparison of Cas12a and Cas9 from various species in vitro revealed that both enzymes share similar types of specificities and tolerate similar mismatches. |
| Knock-in accuracy in mammalian cells | + | ++ | Cas12a mediates efficient and precise endogenous gene tagging via MITI: microhomology-dependent targeted integrations. |
| Key application | Mammalian gene editing | Non-mammalian gene editing and diagnostics | CRISPR/Cas9 system with specific CRISPR-guided nucleases has evolved as prime DNA editing tool in number of gene editing studies in a variety of organisms, including mammalians and primates. The CRIS PR-Cas9 system has been used in cancer therapeutic studies in vivo and ex-vivo. |
The Future of Cas-9 and Cas-12: Beyond Genome Editing
Since the early 2000s, researchers have been trying to engineer new variants to improve the accuracy of CRISPR editing. Cas9-HF1 (or “HiFi”), for example, was designed by J. Keith Joung and team to reduce non-specific contacts 18, making this Cas9 variant particularly useful for research and commercial applications. In parallel, a smaller version of Cas9 was discovered in Staphylococcus aureus, allowing it to be packaged into a single adeno-associated virus (AAV) vector 19. Its small size makes it a potentially viable candidate for therapeutic in-vivo gene transfer. As the first CRISPR effector nuclease, Cas9 has a much longer research history, publication record and greater investment than Cas12.
Interestingly, Cas9 and Cas12 recognize different PAM sequences. The GG sequence required for Cas9 is more abundant in mammalian cells than the canonical TT required by Cas12. For these reasons, many researchers find Cas9 more flexible for use in mammalian systems. In addition, due to its higher specificity and efficiency, CRISPR/Cas9 has been widely applied to many genetic ailments like X-linked diseases, and non-genetic disease such as acquired immunodeficiency syndrome (AIDS), cardiovascular diseases, and neurodegenerative diseases20. Furthermore, researchers are studying the use of the CRISPR/Cas9 technique to cure or alleviate cancers through gene therapy and immunotherapy (ref).
On the other hand, Cas12’s ability to cut single-stranded DNA in a non-specific manner can be leveraged to detect small amounts of DNA from sources such as viruses and cancer cells, making Cas12 a powerful tool for DNA diagnostics. Today, CRISPR tests like those being developed by Dr. Doudna and her team at Berkeley use the various properties of Cas proteins to target illnesses. For example, their diagnostic for HPV uses Cas12a. The test distinguishes between two types of HPV that studies have linked to cervical or anal cancer21.
Conclusion
CRISPR-CasX is a simple system that enables the precise editing of any sequence in the genome of an organism. Today, CRISPR-Cas9 and CRISPR-Cas12 have generated tens of thousands of scientific publications and contribute to the development of novel products and medical advancement ranging from genetically modified crops and livestock to viral therapies. The CRISPR diagnostic technology leverages the different trans-cleavage activities of Cas9, Cas12a (and Cas13) and shows great potential in diagnostic sensitivity, specificity, convenience, and portability22. However, while Cas12 might hold potential for diagnostics applications, it is at a much earlier stage of development than Cas9 and may lack some of the accuracy needed for precision genome editing. Therefore, Cas9 remains the more appropriate option for commercial and research applications where accuracy and reliability are required, particularly in mammalian applications.
References
- Pubmed Search for “CRISPR” – May, 25, 2022. https://pubmed.ncbi.nlm.nih.gov/?term=Crispr&filter=years.2012-2022&timeline=expanded
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429-5433. doi:10.1128/jb.169.12.5429-5433.1987
- Mojica FJM, Juez G, Rodriguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol. 1993;9(3):613-621. doi:10.1111/j.1365-2958.1993.tb01721.x
- Jansen R, Embden JDA van, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565-1575. doi:10.1046/j.1365-2958.2002.02839.x
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174-182. doi:10.1007/s00239-004-0046-3
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiol Read Engl. 2005;151(Pt 3):653-663. doi:10.1099/mic.0.27437-0
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiol Read Engl. 2005;151(Pt 8):2551-2561. doi:10.1099/mic.0.28048-0
- Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol Direct. 2006;1(1):29. doi:10.1186/1745-6150-1-29
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1(1):7. doi:10.1186/1745-6150-1-7
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709-1712. doi:10.1126/science.1138140
- Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18(2):67-83. doi:10.1038/s41579-019-0299-x
- Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi:10.1126/science.aaf5573
- O’Connell MR. Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems. J Mol Biol. 2019;431(1):66-87. doi:10.1016/j.jmb.2018.06.029
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337(6096):816-821. doi:10.1126/science.1225829
- Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a Class 2 CRISPR-Cas system. Cell. 2015;163(3):759-771. doi:10.1016/j.cell.2015.09.038
- Maxmen A. Faster, better, cheaper: the rise of CRISPR in disease detection. Nature. 2019;566(7745):437-437. doi:10.1038/d41586-019-00601-3
- Yan F, Wang W, Zhang J. CRISPR-Cas12 and Cas13: the lesser known siblings of CRISPR-Cas9. Cell Biol Toxicol. 2019;35(6):489-492. doi:10.1007/s10565-019-09489-1
- Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 variants with undetectable genome-wide off-targets. Nature. 2016;529(7587):490-495. doi:10.1038/nature16526
- Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186-191. doi:10.1038/nature14299
- Guo N, Liu JB, Li W, Ma YS, Fu D. The power and the promise of CRISPR/Cas9 genome editing for clinical application with gene therapy. J Adv Res. Published online December 4, 2021. doi:10.1016/j.jare.2021.11.018
- Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436-439. doi:10.1126/science.aar6245
- Kaminski MM, Abudayyeh OO, Gootenberg JS, Zhang F, Collins JJ. CRISPR-based diagnostics. Nat Biomed Eng. 2021;5(7):643-656. doi:10.1038/s41551-021-00760-7
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1(6):e60. doi:10.1371/journal.pcbi.0010060
- Cong L, Ran FA, Cox D, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339(6121):819-823. doi:10.1126/science.1231143
- Mali P, Yang L, Esvelt KM, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339(6121):823-826. doi:10.1126/science.1232033
- Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227-229. doi:10.1038/nbt.2501
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516(7530):263-266. doi:10.1038/nature13769
- Yang L, Güell M, Niu D, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs). Science. 2015;350(6264):1101-1104. doi:10.1126/science.aad1191
- DiCarlo JE, Chavez A, Dietz SL, Esvelt KM, Church GM. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat Biotechnol. 2015;33(12):1250-1255. doi:10.1038/nbt.3412
- Reardon S. First CRISPR clinical trial gets green light from US panel. Nature. Published online June 22, 2016. doi:10.1038/nature.2016.20137
- SHERLOCK team advances its CRISPR-based diagnostic tool. Broad Institute. Published February 15, 2018. Accessed May 15, 2022. https://www.broadinstitute.org/news/sherlock-team-advances-its-crispr-based-diagnostic-tool
- Fogarty NME, McCarthy A, Snijders KE, et al. Genome editing reveals a role for OCT4 in human embryogenesis. Nature. 2017;550(7674):67-73. doi:10.1038/nature24033
- Koo T, Yoon AR, Cho HY, Bae S, Yun CO, Kim JS. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017;45(13):7897-7908. doi:10.1093/nar/gkx490
- Xiao Q, Guo D, Chen S. Application of CRISPR/Cas9-Based Gene Editing in HIV-1/AIDS Therapy. Front Cell Infect Microbiol. 2019;9:69. doi:10.3389/fcimb.2019.00069
- Grunwald HA, Gantz VM, Poplawski G, Xu XRS, Bier E, Cooper KL. Super-Mendelian inheritance mediated by CRISPR–Cas9 in the female mouse germline. Nature. 2019;566(7742):105-109. doi:10.1038/s41586-019-0875-2
- Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149-157. doi:10.1038/s41586-019-1711-4
- Azangou-Khyavy M, Ghasemi M, Khanali J, et al. CRISPR/Cas: From Tumor Gene Editing to T Cell-Based Immunotherapy of Cancer. Front Immunol. 2020;11. Accessed May 25, 2022. https://www.frontiersin.org/article/10.3389/fimmu.2020.02062
- Broughton JP, Deng X, Yu G, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870-874. doi:10.1038/s41587-020-0513-4
- Ledford H. CRISPR treatment inserted directly into the body for first time. Nature. 2020;579(7798):185-185. doi:10.1038/d41586-020-00655-8
- Frangoul H, Altshuler D, Cappellini MD, et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N Engl J Med. 2021;384(3):252-260. doi:10.1056/NEJMoa2031054
- Wang, Dan-Ni. CRISPR/Cas9-based genome editing for the modification of multiple duplications that cause Duchenne muscular dystrophy | Gene Therapy. Published May 9, 2022. Accessed May 25, 2022. https://www.nature.com/articles/s41434-022-00336-3
- Zhang X, Wang J, Cheng Q, Zheng X, Zhao G, Wang J. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 2017;3:17018. doi:10.1038/celldisc.2017.18
- Safari F, Zare K, Negahdaripour M, Barekati-Mowahed M, Ghasemi Y. CRISPR Cpf1 proteins: structure, function and implications for genome editing. Cell Biosci. 2019;9:36. doi:10.1186/s13578-019-0298-7
- Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532(7600):517-521. doi:10.1038/nature17945
- Safari F, Zare K, Negahdaripour M, Barekati-Mowahed M, Ghasemi Y. CRISPR Cpf1 proteins: structure, function and implications for genome editing. Cell Biosci. 2019;9:36. doi:10.1186/s13578-019-0298-7
- Swarts DC, van der Oost J, Jinek M. Structural basis for guide RNA processing and seed-dependent DNA targeting and cleavage by CRISPR-Cas12a. Mol Cell. 2017;66(2):221-233.e4. doi:10.1016/j.molcel.2017.03.016
- Ghorbani A, Hadifar S, Salari R, et al. A short overview of CRISPR-Cas technology and its application in viral disease control. Transgenic Res. 2021;30(3):221-238. doi:10.1007/s11248-021-00247-w
- Dong OX, Yu S, Jain R, et al. Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat Commun. 2020;11(1):1178. doi:10.1038/s41467-020-14981-y
- Belén Paredes M, Eugenia Sulen M. An overview of synthetic biology. Bionatura. 2020;5(1):1088-1092. doi:10.21931/RB/2020.05.01.14
- Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436-439. doi:10.1126/science.aar6245
- Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498-503. doi:10.1038/nature12111
- Kleinstiver BP, Tsai SQ, Prew MS, et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol. 2016;34(8):869-874. doi:10.1038/nbt.3620
- Jones SK, Hawkins JA, Johnson NV, et al. Massively parallel kinetic profiling of natural and engineered CRISPR nucleases. Nat Biotechnol. 2021;39(1):84-93. doi:10.1038/s41587-020-0646-5
- Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 2016;34(8):863-868. doi:10.1038/nbt.3609
- Gao L, Cox DBT, Yan WX, et al. Engineered Cpf1 variants with altered PAM specificities increase genome targeting range. Nat Biotechnol. 2017;35(8):789-792. doi:10.1038/nbt.3900
- Murugan K, Babu K, Sundaresan R, Rajan R, Sashital DG. The revolution continues: Newly discovered systems expand the CRISPR-Cas toolkit. Mol Cell. 2017;68(1):15-25. doi:10.1016/j.molcel.2017.09.007
- Liu X, Homma A, Sayadi J, Yang S, Ohashi J, Takumi T. Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci Rep. 2016;6:19675. doi:10.1038/srep19675
- Bin Moon S, Lee JM, Kang JG, et al. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3′-overhang. Nat Commun. 2018;9(1):3651. doi:10.1038/s41467-018-06129-w
- Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12(4):393-394. doi:10.1016/j.stem.2013.03.006
- Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836-843. doi:10.1016/j.cell.2014.01.027
- Liang P, Xu Y, Zhang X, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363-372. doi:10.1007/s13238-015-0153-5
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230-232. doi:10.1038/nbt.2507
- Wang Q, Alariqi M, Wang F, et al. The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol J. 2020;18(12):2436-2443. doi:10.1111/pbi.13417
- Li Y, Glass Z, Huang M, Chen ZY, Xu Q. Ex Vivo Cell-Based CRISPR/Cas9 Genome Editing for Therapeutic Applications. Biomaterials. 2020;234:119711. doi:10.1016/j.biomaterials.2019.119711
- Mustafa MI, Makhawi AM. SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases. J Clin Microbiol. 2021;59(3):e00745-20. doi:10.1128/JCM.00745-20
- Strecker J, Jones S, Koopal B, et al. Engineering of CRISPR-Cas12b for human genome editing. Nat Commun. 2019;10(1):212. doi:10.1038/s41467-018-08224-4
- Paul B, Montoya G. CRISPR-Cas12a: Functional overview and applications. Biomed J. 2020;43(1):8-17. doi:10.1016/j.bj.2019.10.005
- Wang Z, Zhong C. Cas12c-DETECTOR: A specific and sensitive Cas12c-based DNA detection platform. Int J Biol Macromol. 2021;193:441-449. doi:10.1016/j.ijbiomac.2021.10.167
- Aquino-Jarquin G. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomedicine Nanotechnol Biol Med. 2019;18:428-431. doi:10.1016/j.nano.2019.03.006
- Xiao R, Li Z, Wang S, Han R, Chang L. Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR–Cas12f nuclease. Nucleic Acids Res. 2021;49(7):4120-4128. doi:10.1093/nar/gkab179
- Campa CC, Weisbach NR, Santinha AJ, Incarnato D, Platt RJ. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat Methods. 2019;16(9):887-893. doi:10.1038/s41592-019-0508-6
- Xu R, Qin R, Li H, et al. Generation of targeted mutant rice using a CRISPR‐Cpf1 system. Plant Biotechnol J. 2017;15(6):713-717. doi:10.1111/pbi.12669
- RePORT ⟩ RePORTER. Accessed June 12, 2022. https://reporter.nih.gov/project-details/9776028#details
- CasX start-up Scribe Therapeutics raises $100 million for CRISPR therapies. Chemical & Engineering News. Accessed June 12, 2022. https://cen.acs.org/pharmaceuticals/gene-therapy/CasX-startScribe-Therapeutics-raises-100/99/i12
- Xu X, Chemparathy A, Zeng L, et al. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol Cell. 2021;81(20):4333-4345.e4. doi:10.1016/j.molcel.2021.08.008
- Li Z, Zhang H, Xiao R, Han R, Chang L. Cryo-EM structure of the RNA-guided ribonuclease Cas12g. Nat Chem Biol. 2021;17(4):387-393. doi:10.1038/s41589-020-00721-2
- McGaw C, Garrity AJ, Munoz GZ, et al. Engineered Cas12i2 is a versatile high-efficiency platform for therapeutic genome editing. Nat Commun. 2022;13(1):2833. doi:10.1038/s41467-022-30465-7
- Pausch P, Al-Shayeb B, Bisom-Rapp E, et al. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science. 2020;369(6501):333-337. doi:10.1126/science.abb1400
- 79. Li P, Zhang L, Li Z, Xu C, Du X, Wu S. Cas12a mediates efficient and precise endogenous gene tagging via MITI: microhomology-dependent targeted integrations. Cell Mol Life Sci. 2020;77(19):3875-3884. doi:10.1007/s00018-019-03396-8



