Summary
The CRISPR-Cas9 system has become integral in the toolbelts of genomic scientists, researchers, students, etc. Never has it been so easy to wield a pair of molecular scissors in your (pipetting) hand, however, there are a few intracellular processes to consider before cutting up your genome of interest. In this article, we will give an overview of a few DNA repair pathways that any researcher can take advantage of, and we will discuss when you should, and should not, consider using them. So, whether you are humanizing your gene of interest in a zebrafish model, knocking out a gene in C. elegans, or just want to make a good-old-fashioned fluorescent glowing organism, read on!
Cells come from other cells. We now know that they accomplish this thanks to DNA, which is used as a chemical blueprint to facilitate cells’ growth and reproduction. This being said, it is imperative for the cell/organism to protect its genome. But, sometimes the genome is damaged. In this case, depending on the stage of the cell or nature of the damage, certain DNA damage response (DDR) pathways may be activated Chatterjee & Walker, 2017).
DDR pathways are a collection of intracellular pathways that can sense DNA damage (such as double strand breaks, replication errors, spontaneous deletion, etc.), alert to the presence of damage, and prompt the repair of said damage (Harper & Ellege, 2007). DNA repair pathways can be found in both prokaryotic and eukaryotic organisms and, in fact, many of the DNA repair pathway proteins are highly conserved among all organisms. Different repair pathways exist for different types of DNA damage – damage caused by endogenous reactive oxygen species will have a different mechanism of repair compared to that of exogenously derived damage (like UV or toxic chemical exposure).
While all types of DNA damage are important to recognize and understand, we at InVivo Biosystems are especially interested in double strand breaks (DSBs) repair mechanisms since this is the type of mutation we take advantage of when utilizing the CRISPR-Cas9 system. Contrary to what the average person may believe, the CRISPR-Cas9 system is not directly responsible for the desired genome modification, rather, these molecular scissors generate DSBs and the cell’s endogenous DNA repair pathways do the rest of the heavy lifting.
DSBs are also interesting because they activate two forms of DNA repair: Homology Directed Repair (HDR) and Non-Homologous End Joining (NHEJ). While we are going to focus on HDR and NHEJ for the purpose of this article, there are a number of other DNA repair pathways that exist such as:
- Base excision repair (BER)
- Nucleotide excision repair
- Mismatch repair (MMR),
- Homologous recombination (HR) (Referred to as “homology directed repair/HDR” here)
- Non-homologous end joining (NHEJ)
- A number of Other minor repair pathways
Scientists can take advantage of these endogenous repair pathways to create genetically modified organisms that help further the study of human disease, genetics, basic biology, and more!
Historically, researchers study a complex system by breaking “the system” in some small way and studying how that small break changes the whole. Here at InVivo Biosystems, we utilize the CRISPR-Cas9 (and soon Cas12a) system to induce precisely located double stranded breaks (DSBs) at genes of interest in the zebrafish (Danio rerio) and nematode (Caenorhabditis elegans) genomes. Two of the major DNA repair pathways that are induced when using the Cas9 nuclease are the non-homologous end-joining (NHEJ) and homology-directed repair (HDR) pathways – but when and where is each one used? And what do we need to consider when choosing the mechanism to take advantage of? (that’s right, you have the ability to decide exactly how your cells repair themselves- pretty cool!)
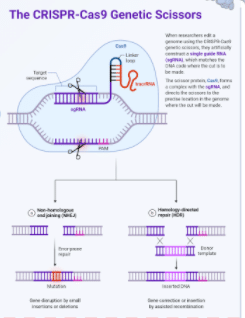
Figure 1. Genetic Scissors (Source: https://biorender.com/nobel-prize-chemistry-2020)
Non-Homologous End Joining (NHEJ)
Non-homologous end joining (NHEJ) is an endogenous DNA repair mechanism that is induced by the creation of double stranded breaks (DSBs) in the genome. It is referred to as “non-homologous” repair because the two broken “ends” of the DNA are indiscriminately rejoined (ligated) back together with minimal reference to DNA sequence. A commonly observed phenomena that accompanies DSBs is the creation of very small single stranded overhangs. These single nucleotide overhangs can create regions of “microhomology” that can help guide DNA repair machinery, sometimes allowing for the perfect repair of the DNA. Unfortunately, this does not occur a majority of the time. Imprecise repair can result in the loss of a small number of nucleotides, effectively knocking out the gene of interest due to INDEL formation resulting in loss of function, frameshift mutations, creation of a premature stop codon, etc. Therefore, this mechanism of DNA repair is best taken advantage of when geneticists want to simply knock out (KO) a gene of interest because this mechanism consistently generates small (1-10 base pair) INDEL errors. To generate these KOs using NHEJ, you will need: Cas9 nuclease (protein or plasmid delivery), single guide RNAs (sgRNA, complexed with Cas9 either in vitro or endogenously via plasmid), and two sets of PCR primers to validate your edit via sequencing.
What if you want to study something a little more complex than a gene knock out? Or maybe you want to study what a slightly modified version of a gene does, or you want to generate a tagged version of your gene of interest? Unfortunately, the NHEJ repair pathway will not allow for more complex genetic modifications such as gene knock-ins (KI). To do this, we must employ the HDR DSB pathway.
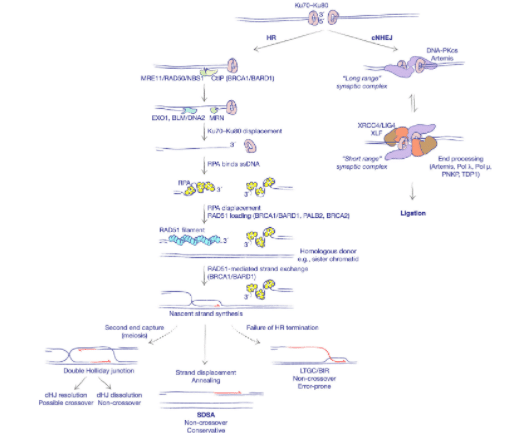
Figure 2. Two major pathways of DNA double-strand break repair (source: Scully 2020, doi:10.1038/s41580-019-0152-0).
Homology-Directed Repair (HDR)
Homology-directed repair (HDR) is an endogenous DNA repair mechanism that utilizes DNA sequence homology to accurately repair DSB damage at the correct genomic location. Unlike NHEJ, which identifies any two broken ends of DNA and “sticks” them back together, the HDR pathway proteins recognize homologous sequences of DNA (from a sister chromatid, a donor homology plasmid, single stranded ODN, etc.) near the region of the DSB and uses those homologous regions as a template for the precise “correction” of the damage (Figure _2_). Researchers can take advantage of this mechanism by flanking any DNA sequence of interest with “homology arms”, or regions of flanking homology to the DSB site. This allows geneticists to effectively insert any sequence of DNA (limitations) into the exact genomic loci as determined by your CRISPR cut sites and flanking homology arms. These inserts are extremely variable and easy to do with HDR: a single AA change at the catalytic site of your enzyme? Done. A fluorescently tagged construct of your gene of interest? Not a problem.
Just because you want to utilize the HDR DNA Damage Response pathway, does not necessarily mean that your organism of choice will utilize it. Sometimes, you have to coax your cells/organism into using the repair pathway that benefits you as the researcher as opposed to the immediate alleviation of DSB repair (benefitting the organism/cell). There are many options for increasing the effective rate of HDR, including, but not limited to: using single-stranded donor homology oligonucleotides, inhibiting NHEJ pathway components (chemically or biologically, i.e. using small interfering RNA (siRNA) or short-hairpin RNAshRNA), genetic modulation of the NHEJ pathway (i.e. using NHEJ to knock out NHEJ before you use HDR to edit your organism), and cell cycle synchronization (HDR is typically restricted to the S and G2 phases)..
Some design considerations to consider include placement of your homology arms which should be as close to the DSB site as possible. You also need to consider the size of your arms, Liu et al. found that 50-100% of the size of your insert seemed to work well.. For more considerations, you can check out the work of Liu et al. and Yang.
Conclusion
In summary, researchers can utilize HDR to create unique KO/KI constructs/ tagged KI inserts/ etc., and, unlike NHEJ, is not only limited to KOs. The major difference when it comes to the reagents involved with NHEJ vs HDR is the presence of some DNA with regions of homology that correspond to the flanking regions of the DSB site. To the researchers and geneticists, this means including either a plasmid or single stranded DNA with your insert flanked by regions of homology corresponding to the DSB site you are inducing with Cas9 in your CRISPR-Cas9 mix.
To learn more about how Cas9 is used checkout our other blogs: https://invivobiosystems.com/view-from-the-bench/zebrafish-genome-editing-crispr-vs-tol2/, https://invivobiosystems.com/crispr/the-state-of-knock-ins-in-zebrafish/
Are you looking for a reliable and efficient way to introduce specific genetic changes into your target cells for your research? Our CRISPR Knockin service might be the solution you’re looking for! Our team of experts has the knowledge and expertise to help you achieve your research goals using the latest technology available. Contact us today to learn more about our services and how we can help you with your research needs.
References:
Chatterjee, N., & Walker, G. C. (2017). Mechanisms of DNA damage, repair, and mutagenesis. Environmental and molecular mutagenesis, 58(5), 235-263. https://doi.org/10.1002/em.22087
Liu, M., Rehman, S., Tang, X., Gu, K., Fan, Q., Chen, D. and Ma, W., 2019. Methodologies for Improving HDR Efficiency. Frontiers in Genetics, 9.



