Summary:
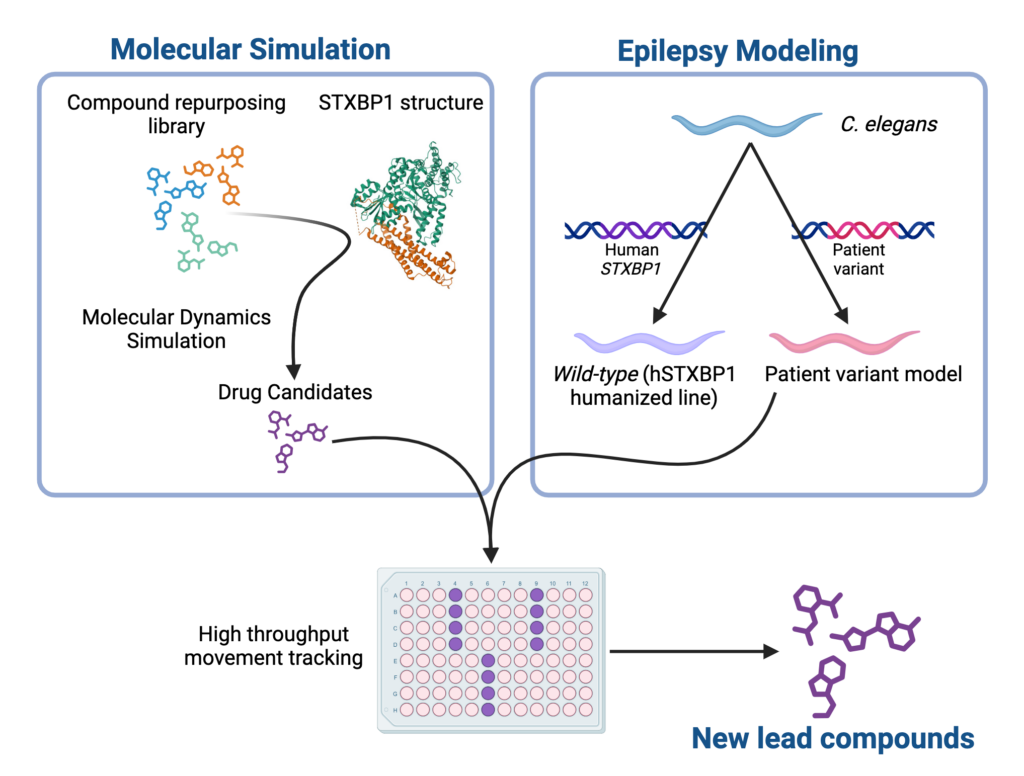
Computational modeling of STXBP1 protein docking with a compound repurposing library revealed a list of 15 drug candidates.
Candidates were testing using a validated C. elegans model of epilepsy carrying a humanized allele of STXBP1.
Study returned 3 compound “hits” that partially reversed the variant phenotype, including one already used for treatment.
Confronting the Reality of “No Cure”
Hearing the words “There is no cure” is a devastating blow for patients and parents confronted with the diagnosis of a rare disease. The journey for individuals grappling with rare diseases is marked by the struggle to secure accurate diagnoses and, once diagnosed, a scarcity of viable treatment options. Yet, amid these challenges, the evolving technological landscape is revealing hope along the horizon. In this post, we highlight a case study of how small animal models are opening new doors for those facing the complexities of STXBP1, a rare form of epilepsy.
The Enigma of STXBP1
Epilepsy, a neurological disorder affecting over 65 million people worldwide, is characterized by recurrent seizures stemming from abnormal electrical activity in the brain. Within the spectrum of epilepsy, STXBP1 stands out as a rare variant caused by mutations in the gene encoding the STXBP1 protein. Unfortunately, individuals with STXBP1 epilepsy encounter limited treatment options compared to other forms of epilepsy. Organizations such as the STXBP1 Foundation and the Rare Smile Foundation have emerged to support research and to fill the gap left by mainstream pharmaceutical companies that focus on therapies with broad demand.
Beyond the Lock and Key: Evolution of Drug Development
Traditionally, drug development involves screening vast libraries of chemical compounds to find those capable of reversing a condition, often without specificity to the protein target. However, recognizing the cost and time associated with such endeavors, scientists are turning to computational modeling to identify chemical keys that precisely fit the proteins involved. This approach, known to chemists as Molecular Dynamics (MD), simulates the movement of individual atoms in space. When applied to proteins in solution, it is extremely computationally intensive, but advances in methodology and AI are making the approach more accessible.
In collaboration with Miguel Weil and QR Genetics, we used MD to simulate the docking of chemicals in a compound repurposing library to the structure of the STXBP1 protein. This identified those compounds from the library with the right shapes and functional groups to possibly interact directly with STXBP1. By leveraging this technology, research groups with budget constraints can reduce a library of thousands of compounds down to that handful most likely to interact with the target protein, and thus decimate the time and expense of the drug screening process.
Nematode Navigators: The Surprising Prowess of C. elegans
The next challenge was to test the efficacy of these compounds in relieving the symptoms of STXBP1 disorder. Creating and testing rodent models for rare diseases is costly and time-consuming enough in the mainstream pharmaceutical industry, but prohibitively expensive for many rare disease organizations needing fast answers for a small community. Enter the nematode worm, C. elegans, a tiny organism that plays a colossal role in our research.
The speed and scalability of the C. elegans model were demonstrated in our previous work in which we quickly modeled over ninety patient variants of the STXBP1 gene to test their pathogenicity. These worms had been “humanized” by replacing the othologous nematode gene unc-18 with a copy of human STXBP1. The utility of this model lies in the fact that the human STXBP1 gene reverses the paralysis caused by unc-18 loss of function. [link to previous blogs] (We have also extended our modeling of STXBP1 disorder to Zebrafish [link]).
For this study, we leveraged our validated humanized STXBP1 line, introducing a patient-specific mutation, R406H, that causes a specific paralysis phenotype that is the worm equivalent to epilepsy. These mutant worms were exposed to a total of 15 drugs selected from the preliminary MD simulations, and their movements recorded over time in a high-throughput movement analysis platform. Remarkably, three of these drugs induced a significant reduction of the paralysis caused by the R406H mutation, compared with the “wild-type” STXBP1 control. This finding exemplifies the power of small animal models in facilitating rapid and cost-effective preclinical testing.
A Fortuitous Connection
But would these drugs have a therapeutic effect in humans? The extraordinary aspect of this study lies in the revelation that a medication that some patients are already taking had been added to the group of 15 test compounds and blinded to the researchers. It turns out that his drug was among the three hits that had a positive effect on movement! This not only validates the study but also instills confidence in the therapeutic potential of the other identified drug hits.



