Polymerase chain reaction (PCR) is a robust and familiar technique for genotyping in laboratories around the world for over thirty years. The PCR method is used to create millions of copies of a specific, targeted, fragment of DNA which can then be studied in closer detail. Since 2009, this tried and true technology has been at the core of the genotypic screening process of transgenic organisms created here at InVivo Biosystems.
When used as part of the screening method to det
ect precise deletions (knockout: KO) or insertions
(knock-in: KI) in the genome of C. elegans, the PCR method provides consistent and reliable results. The DNA products created from PCR are frequently analyzed using gel electrophoresis. When these products are run on an agarose gel, the differences in fragment size between mutant and wildtype organisms (as typically seen in KO/KI builds) can be easily visualized. This allows laboratory technicians to quickly and confidently identify potential transgenic candidates for sequencing confirmation of the desired edit.
Unfortunately, using some of these traditional methods to screen for smaller genomic changes, such as point mutations, can be labor intensive, time consuming, and fraught with inconclusive results (such as false positives and/or negatives). When screening point mutation builds, the resulting PCR product may be the same size for both mutant and wildtype strains. It is no longer p
ossible to differentiate between two different animals by comparison of PCR product size with gel electrophoresis. It is often necessary to employ other, less robust, screening techniques to identify the desired genotypic edit. Allele specific PCR and restriction enzyme digestion are two common alternative screening methods. And while they are proven to work, these tests are often unreliable with difficult to interpret results. This has the potential to cause unnecessary rework, and can delay completion and increase delivery time of the desired transgenic.
Is there a better way to screen transgenic builds involving small genomic mutations? Perhaps something faster, more robust, and that provides more reli
able results?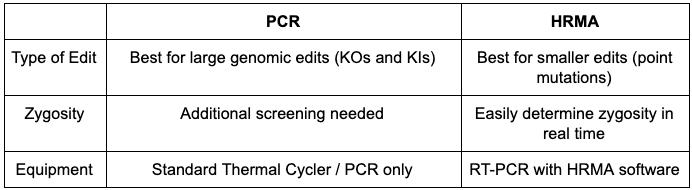
High Resolution Melt Analysis (HRMA), in contrast, excels at genotypic screening processes for transgenic builds involving point mutations. HRMA is a powerful screening tool that allows for the differentiation between mutant and wildtype PCR products based on their own unique and distinct melting profiles. To perform these assays, we use a real-time PCR detection system and dedicated HRMA data collection software. This allows laboratory staff to monitor the results in real-time as the assay is being performed. This is just one of the many advantages to this screening method.
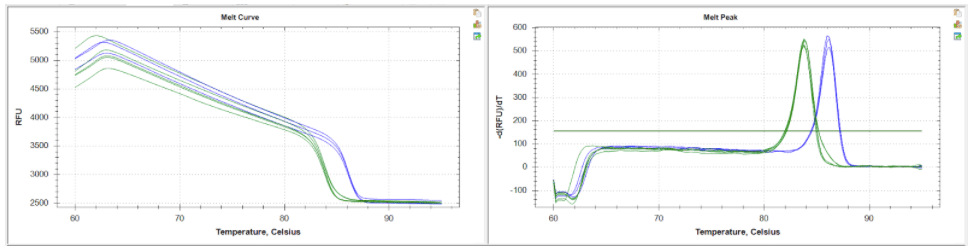
Figure 1: HRMA melt curve and melt peak traces as shown using BioRad CFX Maestro software. Homozygous mutant candidates (green trace) with a predicted melting point of 82.5°C as compared to wildtype control samples (blue trace) with a predicted melting point of 86.5°C. Candidates of interest are easily differentiated and identified in real time.
Similar to traditional screening, PCR amplification is the first step of the process. However, a fluorescent dye is added to the reaction mix that binds to the DNA product created by the PCR reaction. The resulting PCR product is then precisely heated in small increments, causing dissociation of the DNA fragments. The HRMA software monitors the dissociation and subsequent release of the fluorescent molecules. Variations in the nucleic acid sequences (such as point mutations) between mutant (homozygous/heterozygous) and wildtype fragments will result in distinct melt curves displayed by the software (Figure 1). This allows for easy identification of potential transgenic candidates without the need for time consuming and less reliable screening methods. Additionally, because the HRMA process is not damaging to the DNA product, it is also possible to use the resulting product for sequencing purposes without the need to run another PCR reaction!
With all of these advantages over traditional screen methods, HRMA screening allows us to quickly and confidently detect and identify the desired point mutations in our transgenic animal builds. Use of this cutting edge genotyping technology and user friendly analysis allows InVivo to consistently build and deliver accurate transgenic animals to clients for all of their research purposes.
InVivo Biosystems offers a variety of C. elegans services packages to fit the needs and budget of every lab. Learn more.

