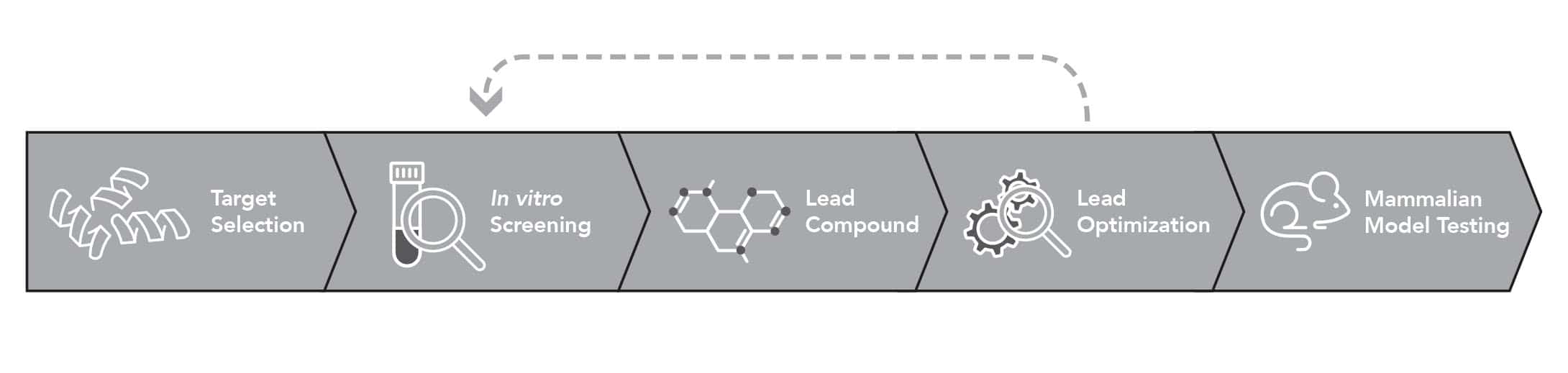
History of Drug Discovery
With the advent of the molecular biology revolution and the human genome project in the 1990s there was a historic shift toward target-based drug development. Target-based drug development, or TDD, relies on prior knowledge of disease mechanisms and targets before development can begin.
This change coincided with a loss of productivity in identifying new drugs. In 2011, Swinney and Anthony released a report that provided a strong indictment of the target-based approach to drug discovery: between 1999 and 2008, only 37% of the first-in-class small molecule drugs approved during this time came from target-based screens (Swinney and Anthony 2011). The remaining were identified using phenotype-based drug discovery (PDD), in which molecules are evaluated for their ability to ameliorate disease phenotypes, rather than interaction with known disease targets. Phenotype-based drug discovery, in which drugs are tested for their efficacy on disease phenotypes rather than specific targets, appears to be significantly better at identifying new drugs.
This finding has resulted in a pendulum swing back towards phenotypic discovery – and in the past decade, first-in-class drugs for Spinal Muscular Atrophy (SMA), Hepatitis C virus, and cystic fibrosis (CF) were all identified via this method (Swinney and Lee 2020). However, in order to fulfil the promise of PDD, careful consideration of the assay system and phenotypic expression is needed.
Target-based vs. Phenotype-based Drug Discovery
Considerations for PDD
Vincent et al have proposed a “rule of 3” to increase the success of phenotypic screening (Vincent et al. 2015). These rules state that increasing similarity between the human presentation of a disease and the phenotypic screen provides a higher chance of translational success, because the premise of the “unbiased search” of phenotypic screening – namely, that biology is accurately recapitulated so that compounds can be identified by their efficacy, rather than by engagement with a known target – is upheld. Practically, this is broken down into three areas: the assay system, the method of disease induction, and the phenotypic readout.

RULE 1: The Assay System must be similar in genetics and/or complexity.
Assay systems for PDD cover a broad range of available models, from workhorse immortal cell lines (e.g. HEK293 cells) to patient-derived iPSCs to transgenic model organisms. A clear gradient exists whereby increasing either the genetic similarity or the tissue complexity and embodiment of the assay system also increases the translatability of the results obtained. Genetic models of disease pathways can range from simple (e.g., overexpression assays using a single gene, uncontrolled copy numbers and integration sites) to the sophisticated (e.g., single-copy insertions of multiple disease-related genes at sites with known genomic regulation, or modeling of particular single nucleotide polymorphisms (SNPs) or splice variants). Perhaps the best genetic model of a human disease is found in induced pluripotent stem cells, or iPSCs, wherein all the genetics represented in a specific human are maintained to represent disease. However, this approach is costly, and ultimately the results may not generalize because any single human may have interacting background mutations that are difficult to identify post-hoc. Additionally, iPSCs suffer from deficits in tissue complexity and embodiment.
Another factor to consider in choosing an assay system is overall complexity. 2D, single cell type systems are commonly used and easy to maintain, but suffer in translatability because they cannot model organism-level regulation through interaction of multiple cell types, organ systems, or endocrine factors. 3D cell culture and organoids improve on this situation by providing accurate representations and organizations of multiple cell types, and yet, in many cases they are limited to the superficial layer of the organ system and again cannot model any organism-level regulation of disease manifestation. In many cases, cells are a poor substitute for organisms, and the best models of human disease are in vivo systems. Rats, mice, fish, and worms have all been used for this purpose. Rats and mice have obvious advantages in genetic similarity and embodiment, however, modern genetics, including the CRISPR revolution, is starting to change the landscape because human genes can now be expressed, manipulated, and visualized in small animal models such as worms and fish. This provides increased genetic relevance while maintaining the benefits of tissue embodiment and cost effectiveness.
![]()
RULE 2: Genetic or biological methods of inducing the Disease State are better than artificial methods.
Many methods can be used to induce cells or organisms into a disease state where phenotypes are expressed. Here again, a high degree of similarity to the manifestation of disease in humans is likely to give a more translatable result. Low-complexity, low-relevance stimuli such as chemical perturbations are unlikely to yield meaningful results, because they may use different biological mechanisms to induce disease phenotypes. Biological or multi-pronged stimuli, such as RNAi or cytokine cocktails, are a better option. However, the most relevant disease manifestation is one in which no exogenous stimulus is required. In this case, the genetic perturbations are sufficient to induce the disease state in the model system. This method utilizes the endogenous biological pathways to produce phenotypic perturbations of disease.
RULE 3: The Phenotypic Readout should be representative of the disease.
Identification of drugs that can truly ameliorate disease presentations will be most relevant when the phenotypic readout is representative of the disease (e.g., using muscle function as a readout of ALS). Thus, care must be taken to ensure that phenotypic readouts are close to the disease state itself, rather than abstracted readouts such as single gene or protein expression. Although they facilitate screening, such endpoints can mislead hit discovery because they are too limited and myopic as a representation of overall organismal function.
Conclusions
The “rule of three” proposed by Vincent et al provides a practical framework for the design of phenotypic drug discovery programs that improve the efficiency of the pharmaceutical industry. Although there are tradeoffs between relevance and expense and effort, advances in genetic engineering are increasing access to high-quality models of human genetics in small-animal model organisms that are typically overlooked. These models may make a good choice as a screening tool because they have a high degree of tissue complexity and can represent organism-level responses. Additionally, they have disease-associated functional phenotypes that can be mapped easily onto human expressions of disease.
References
- Swinney, David C., and Jason Anthony. 2011. “How Were New Medicines Discovered?” Nature Reviews Drug Discovery. https://doi.org/10.1038/nrd3480.
- Swinney, David C., and Jonathan A. Lee. 2020. “Recent Advances in Phenotypic Drug Discovery.” F1000Research 9 (August). https://doi.org/10.12688/f1000research.25813.1.
- Vincent, Fabien, Paula Loria, Marko Pregel, Robert Stanton, Linda Kitching, Karl Nocka, Regis Doyonnas, et al. 2015. “Developing Predictive Assays: The Phenotypic Screening ‘rule of 3.’” Science Translational Medicine. https://doi.org/10.1126/scitranslmed.aab1201.
InVivo Biosystems’ drug discovery development services are designed to propel your research forward and revolutionize the future of medicine.