The right CRISPR Knock-In zebrafish line–a fluorescent tag for following protein activity, or a precise nucleotide change for modeling disease–can be a game changer for a researcher. CRISPR technology has made creating Knock-Ins much easier, but challenging hurdles still remain. Many studies have examined different techniques and how several parameters impact Knock-In success; however, there is not a consensus on which parameters are generally ideal for generating knock-ins. In this post we share a few key considerations that have enabled successful Knock-Ins for our clients and highlight a customer’s successful creation of a fluorescently-tagged zebrafish line.
Key Points
- Creating CRISPR Knock-Ins in Zebrafish can be challenging, but emerging tools offer ways to improve success.
- One key consideration is choosing the correct form of donor homology template.
- Our success and that of our clients have help uncover some key parameters to improve the outcome of Knock-In edits.
CRISPR Knock-Ins: What type of edit is your goal?
When we say “Knock-in,” we are referring to using a CRISPR/Cas9 system to create a precise genome edit by cutting the DNA at a defined site then integrating a new sequence during repair. This method typically depends on the process of homology driven repair (HDR) to introduce a precise repair with an exogenously supplied DNA Template. First, one or more single guide RNAs (sgRNA) direct Cas9 nuclease to make precise cuts in the genome at the location of the desired edit. The sequence of interest, flanked by homology arms (sequences complementary to the regions surrounding the edit) then acts as a repair template, replacing the native sequence with an engineered version. The inserted construct may include or consist of a tag, a protein marker designed to allow reliable detection of the protein of interest.
There are many considerations for using the KI system including biological obstacles that may interfere with the success of knocking-in sequences. In terms of biological obstacles we must consider the two prominent forms of DNA repair present during a Knock-In: homology-driven repair (HDR) and non-homologous end joining repair (NHEJ). Zebrafish embryos develop relatively rapidly with highly proliferative cells, limiting the efficiency of HDR. While HDR efficiency is limited in this model, NHEJ is much more dominant when cells are rapidly dividing such as in early embryos. NHEJ is an efficient repair mechanism but the DNA strands are indiscriminately rejoined with minimal reference to the sequence, unlike HDR which repairs DNA sequences accurately and in the correct genomic location. The prominence of NHEJ makes it more likely that your template was not properly or completely knocked-in, an inherent challenge to KI projects.
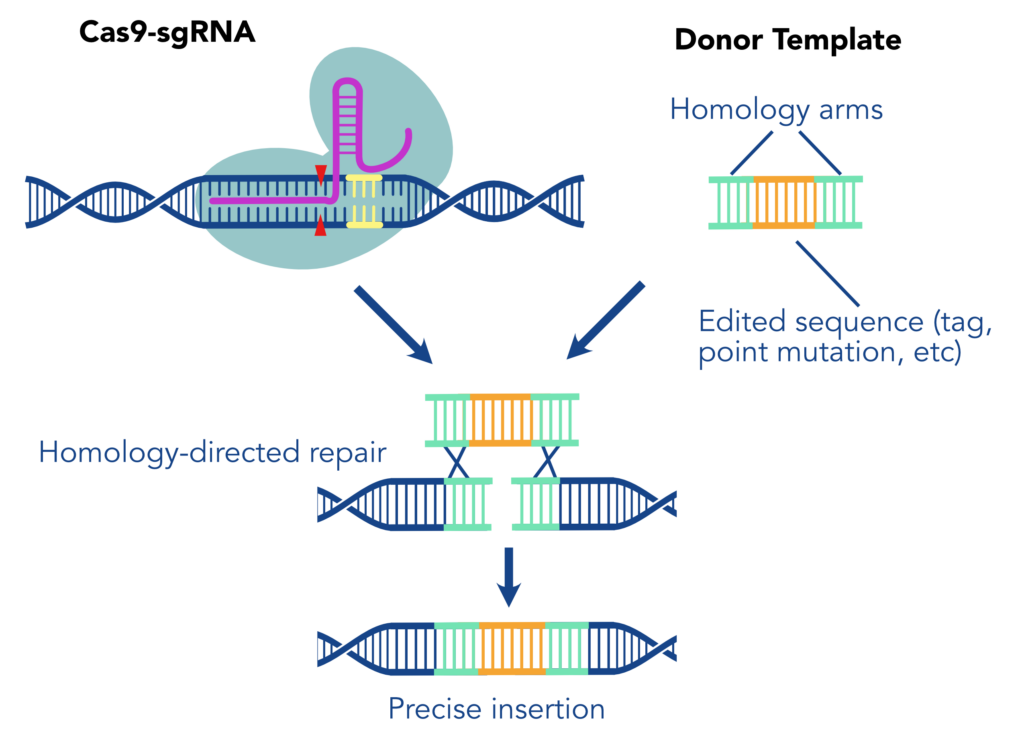
Choosing the Right Template
- Which type of repair template to use - single or double stranded DNA, or viral vectors?
- Homology arm organization and specifications - length and symmetry around edit.
- Stranding (e.g. sense vs anti-sense) for single stranded templates.
Other challenges faced when completing a KI include the type of template to introduce the desired edit. Currently there are two relevant types of repair templates receiving widespread use in knock-in editing including: double-stranded DNA (dsDNA) or plasmid-based templates, short single-stranded DNA (ssDNA). Both strategies are utilized to generate stable germline transgenic zebrafish, which is InVivo Biosystems’ specialty. A significant sticking point in KIs is the size of the edit; smaller edits have proven to be more attainable in the context of KIs than a few hundred base pairs, especially given the efficiency of NHEJ during zebrafish development. For genome edits greater than several hundred base pairs (bp), dsDNA templates are primarily used (Table 1). At this size, production of ssDNA templates can be time-consuming with limited yields using current methodologies or prohibitively expensive and require ordering from dedicated manufacturers to ensure high quality, error-free templates (Iyer et al., 2019). However, plasmid-based templates can result in random, off-target insertion of plasmid DNA into the host genome and have been found to possess higher rates of cytotoxicity than ssDNA template counterparts. Despite the lower cargo capacity (on average <200bp), ssDNA templates (specifically single-strand oligodeoxynucleotides, or ssODNs) have demonstrated a greater efficiency in the HDR repair pathway in addition to lower cytotoxicity (Biswas & Enzmann 2021). Synthetic repair templates such as ssODNs have also demonstrated a lower rate of the off-target integration events found in the use of plasmid-based templates (Boel et al 2018).
Here at InVivo Biosystems we are exploring how template design can improve knock-in efficiency and have seen great success with introducing point mutations and small cargo (<150 bps) such as loxP cassettes or 3xFlag tag epitopes into zebrafish, which are especially challenging for KI experiments due to their rapid cell cycling in early embryos. Given the large variety of genes and loci we are targeting for our clients, we are also starting to identify some of the key design principles that maximize the chances for successful knock-in for our clients.
How can we help with your Knock-In project?
Additional Design Considerations
Given the large variety of genes we are targeting for our clients, we are also starting to identify some of the key design principles that maximize the chances for successful knock-in for our clients. The best way to ensure a smooth start to KI projects is to start with the most ideal reagents (sgRNAs, primers, etc). During the sgRNA selection process important points to consider include the location of the sgRNA, cutting efficiency, and binding sites in relation to the edit of interest. We can also utilize unique features of the loci like single nucleotide polymorphisms (SNPs), indels, and duplications to create primers that are specific enough to amplify our region of interest. Once reagents have been validated both phenotypic and genotypic screenings are implemented to confirm germline transmission, taking advantage of fluorescent tags to help confirm potential founder lines transmitting the desired edit. InVivo Biosystems have the capabilities to validate reagents and screening methods to overcome the challenges posed by KI projects.
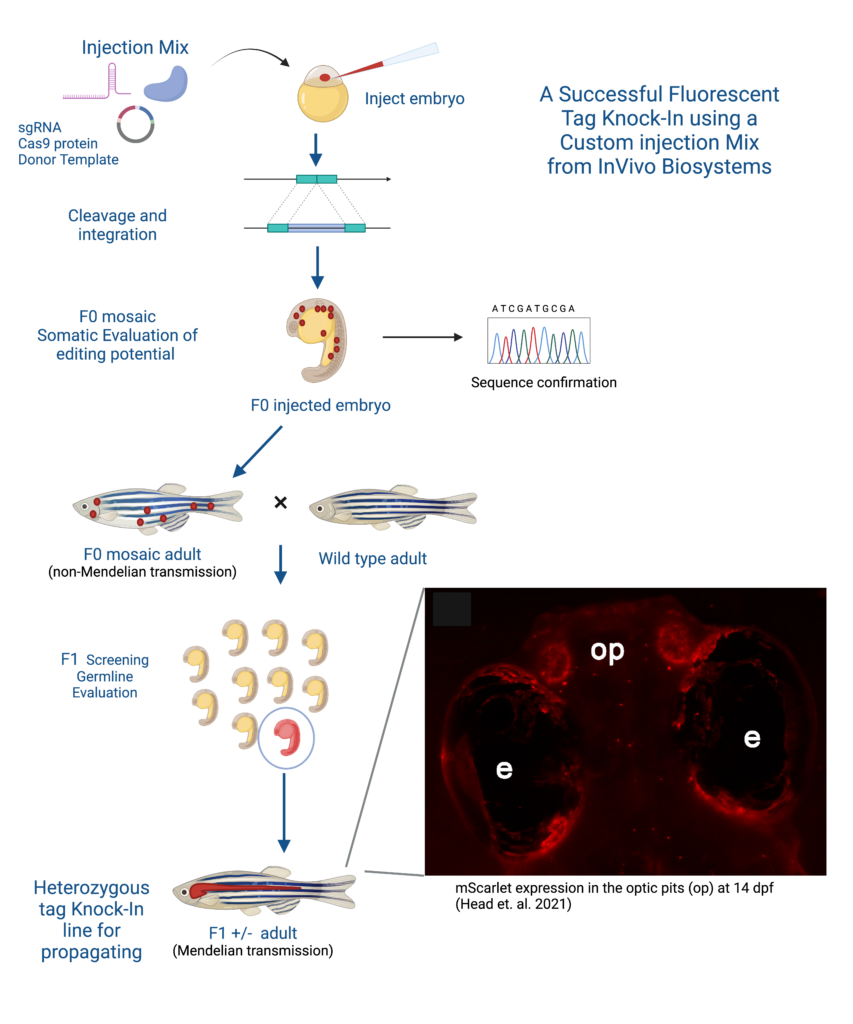
Customer Story
A successful example of generating a stable D. rerio line using the KI strategy is the collaboration between Dr. Maret Traber, a distinguished professor at Oregon State University, and InVivo Biosystems. The Traber Lab has done extensive research on the effect of Vitamin E, and has shown Vitamin E deficiency is linked to energy metabolism deregulation, gene expression networks, and nervous system development but needed a more reliable way to track Vitamin E and proteins in zebrafish models (McDougall et al., 2017; Head et al., 2021b; Miller et al., 2021; Head et al., 2020). By partnering with InVivo Biosystems the Traber Lab was able to create a new fluorescent-tagged zebrafish line to study the effects of Vitamin E on embryonic development by strategically knocking-in the red fluorophore mScarlet generating a Ttpa-tagged zebrafish model.
Conclusion
Dr. Traber’s story is just one of the several examples in which we here at IVB have harnessed the power of CRISPR technology to precisely introduce desired genetic modifications in zebrafish, revolutionizing studies in developmental biology, disease modeling, and beyond. Our experienced team of scientists is ready to assist you in generating custom knock-in zebrafish lines tailored to your specific research goals. Take your research to the next level with our Zebrafish Knock-in service!
References
Biswas & Enzmann (2021). Optimizing CRISPR Knock-ins: Tips and Tricks For Successful Knock-in Editing, https://www.synthego.com/blog/crispr-knockin-tips-tricks
Annekatrien Boel, Hanna De Saffel, Wouter Steyaert, Bert Callewaert, Anne De Paepe, Paul J. Coucke, Andy Willaert; CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis Model Mech 1 October 2018; 11 (10): dmm035352. doi: https://doi.org/10.1242/dmm.035352
Evans, H. M., & Bishop, K. S. (1922). ON THE EXISTENCE OF A HITHERTO UNRECOGNIZED DIETARY FACTOR ESSENTIAL FOR REPRODUCTION. Science (New York, N.Y.), 56(1458), 650-651. https://doi.org/10.1126/science.56.1458.650
Gaj, T., Staahl, B.T., Rodrigues, G., Limsirichai, P., Ekman, F., Doudna, J.A., Schaffer, D.V., (2017). Targeted gene knock-in by homology-directed genome editing using Cas9 ribonucleoprotein and AAV donor delivery, Nucleic Acids Research, 45 (11), Pg. e98, https://doi.org/10.1093/nar/gkx154
Head B, La Du J, Barton C, Zhang J, Wong C, Ho E, Tanguay RL, Traber MG. (2021a) RedEfish: Generation of the Polycistronic mScarlet: GSG-T2A: Ttpa Zebrafish Line. Antioxidants; 10(6):965. https://doi.org/10.3390/antiox10060965
Head, B., Ramsey, S. A., Kioussi, C., Tanguay, R. L., & Traber, M. G. (2021b). Vitamin E Deficiency Disrupts Gene Expression Networks during Zebrafish Development. Nutrients, 13(2), 468. https://doi.org/10.3390/nu13020468
Head, B., La Du, J., Tanguay, R.L. et al. Vitamin E is necessary for zebrafish nervous system development. Sci Rep 10, 15028 (2020). https://doi.org/10.1038/s41598-020-71760-x
McDougall, M., Choi, J., Kim, H. K., Bobe, G., Stevens, J. F., Cadenas, E., Tanguay, R., & Traber, M. G. (2017). Lethal dysregulation of energy metabolism during embryonic vitamin E deficiency. Free radical biology & medicine, 104, 324-332. https://doi.org/10.1016/j.freeradbiomed.2017.01.020
Iyer et al. (2019). Efficient Homology-directed Repair with Circular ssDNA Donors, http://dx.doi.org/10.1101/864199
Miller, G. W., Labut, E. M., Lebold, K. M., Floeter, A., Tanguay, R. L., & Traber, M. G. (2012). Zebrafish (Danio rerio) fed vitamin E-deficient diets produce embryos with increased morphologic abnormalities and mortality. The Journal of nutritional biochemistry, 23(5), 478-486. https://doi.org/10.1016/j.jnutbio.2011.02.002
Shah, R. S., Rajalakshmi, R., Bhatt, R. V., Hazra, M. N., Patel, B. C., Swamy, N. B., & Patel, T. V. (1987). Vitamin E status of the newborn in relation to gestational age, birth weight and maternal vitamin E status. The British journal of nutrition, 58(2), 191-198. https://doi.org/10.1079/bjn19870086




