A great article that highlights some of the strengths of C. elegans for investigating higher level processes beyond what can be easily studied using cell culture. In this case alternative pre-mRNA splicing. Also helped expose me to a new descriptor for C. elegans worms – intron-rich.
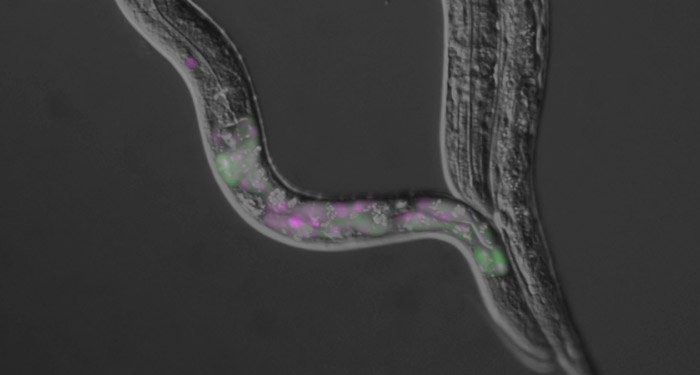
A review article in WIREs RNA by Wani and Kuroyanagi from Tokyo Medical and Dental University summarizes advantages of the model organism Caenorhabditis elegans in elucidating principles of alternative pre-mRNA processing regulation in multicellular organisms. The tiny nematode worm C. elegans has just 1,000 cells, yet is comprised of multiple tissues such as the nervous system, muscles, digestive tract, and hypodermis. Despite its small number of cells, C. elegans has about 20,000 protein-coding genes, up to 25% of which undergo alternative pre-mRNA processing. What makes C. elegans ideal for studying mRNA processing among model organisms is that it is transparent. Use of multiple fluorescent proteins as a readout of cell-type-specific pre-mRNA processing enables searching for factors and genomic sequences necessary for the regulation in living worms.
According to the review article, the regulation factors and sequences discovered thus far in C. elegans turned out to be highly similar to those in mammals. Future studies with this animal will provide further insights into the regulation and evolution of critical alternative pre-mRNA processing events.