Summary
The human microbiome is a booming area of research thanks to evidence showing that it could have far-reaching effects on our health. But how is this research being conducted? In this article (a subsection of our Microbiome White Paper), we will discuss the commonly used model organisms for microbiome research, and what insights they can provide.
Mice Models
While mice models have predominantly been used in microbiome research, over the years researchers have pointed out aspects of the model which make it less than ideal for researching the microbiota of humans. Beyond the standard challenge of murine models (high maintenance costs, and long life span) the ability of mice to be a translatable model for human microbiota research is also limited due to structural differences in intestinal tracts. There are also major differences between the two species’ intestinal microbiota. Studies have shown that only 4% of microbial genes in mice and humans share considerable identity, and the quantity of the shared bacterial types vastly differ [1]. Furthermore, the validity and reproducibility of mouse microbiota studies have been questioned, as rearing facilities (particularly the feed) and genetic backgrounds have significant impacts on microbial composition and multiple different mouse strains have been used [2,1,3]. Finally, it has been found that studies often lack comparable chow (mouse food): in 2007 only 14% of papers regarding diet used identical feed [2,1,3]. These drawbacks have led microbiologists to “wonder why mouse models are used so often for translation to humans and the simple answer could be that there is no better alternative” [1] – but alternative models are starting to emerge!
Human vs Mice Microbiome
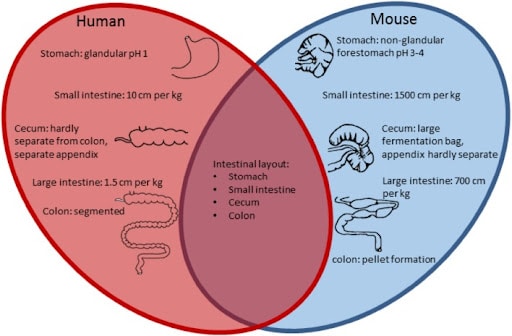
Figure 1. Comparison of the intestinal tract features of human and mouse – the main similarities and differences displayed as a Venn diagram [43]
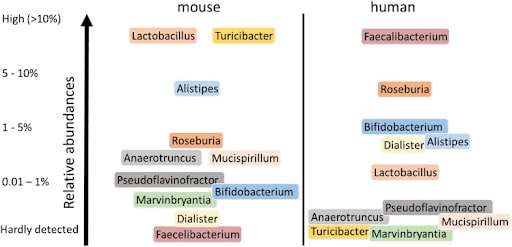
Figure 2. Major different human and murine intestinal genera. Only genera are shown that showed consistent differences in relative abundance between humans and mice [1].
Alternative Systems for Modeling the Human Microbiome
Computational Models
Computational models are useful for insight into the effects of microbes in humans without having to consider interspecies differences. Yet a computational approach is currently limited, because complete models of the microbiota-gut-axis do not exist [4]. It is likely computational modeling will become a powerful tool as a richer and more complete understanding of the microbiome interactions is generated.
Zebrafish Models
Zebrafish are rapidly becoming a popular tool in microbiome research. Their fast development, small size, and high similarity with the human genome (70% of human genes have fish orthologues), makes them an advantageous model [5]. Their ex-utero development, and transparent embryos, make observational studies of host-microbe systems easy. Additionally, zebrafish embryos can be sterilized and then grown microbe-free vs chosen microbes for direct examination of microbe-host interactions [6]. Furthermore, what may appear to be a limitation, their aquatic environment, is actually beneficial for microbiome research as it provides a controllable environment, and their ability to be fed the same diet their whole lives removes the confounding factor of diet in microbiome research [5]. Zebrafish research also benefits from established techniques for microscopy, chemical screening, and genome editing. While the zebrafish does not have a stomach, its intestine is highly homologous to the mammalian intestine, and it has been shown that the regulatory mechanisms controlling the intestinal system are highly conserved between zebrafish, mice, and humans [7,8].
Despite being a relatively new model for investigating microbiome mechanisms, zebrafish research has provided compelling insights: for instance, they have illustrated how impactful interhost dispersal of microorganisms can be to the variation seen in microbiomes. When wild-type zebrafish and immune-deficient zebrafish (myd88 knockouts) were housed separately, their microbiome composition was significantly different, however, when they were housed together these differences were largely eliminated [9]. Zebrafish have also been used to investigate diet’s effect on microbiome – showing that microbiota regulate intestinal absorption and metabolism of fatty acids in zebrafish, and that microbiota assembly can be influenced by a high versus low-fat diet [5,10]. Like humans, there is interindividual variation in zebrafishes’ intestinal microbiota. And developmental-microbiome studies have revealed that their microbiome follow a similar trajectory to humans (low to high complexity; age specific populations, etc). Intriguingly, as occurs in humans [11], the diversity of their microbiome declines with age [12].
Zebrafish are a particularly good model to study the effect of exogenous factors such as toxins and pathogens on the microbiome since zebrafish are widely used in toxicology research. These studies have shown that widespread environmental chemicals disrupt microbiota early in life, and that parasites can restructure the gut microbiome’s composition [13,14]. Consequently, microbiome profiling could be used as an early way to diagnose infection or bacterial agents that will drive disease progression as people age.
The bidirectional signaling cascade of the microbiota-gut-brain axis (MGBA) is well-established, but more recently dysbiosis has been implicated in neurological disorders such as autism spectrum disorder and Alzhiemer’s disease [15]. Zebrafish have emerged as a promising model to better understand MGBA, due to the conserved features of the GIT between zebrafish and humans, the newly generated zebrafish models of autism spectrum disorder and Alzheimer’s disease, and the similarity between zebrafish and humans’ enteric nervous system (ENS) [15]. In both species, the ENS regulates intestinal motility and communication between the central nervous system and the intestine using many of the same neurotransmitters [16]. Furthermore, zebrafish and humans have many of the same immune-signaling pathways, which have been implicated in neurodegenerative diseases.
“With its ability to be utilized in many different areas of microbiome research, zebrafish are surely going to continue contributing to this field.“
C. elegans Models
The nematode, C. elegans has become a premiere model for microbiome research; a simple multicellular organism, C. elegans are easy to maintain, have a short lifespan, transparent body throughout its life and it is amenable to high-throughput studies (entire lifecycle an be examined in 1536 microwell plates) [17]. There are also many morphological and functional similarities between C. elegans and the human gut which makes this simple animal a highly translatable model to humans. Notably, many metabolic, stress, and innate immune pathways implicated in regulating microbiome recruitment are conserved between humans and C. elegans [18]. The C. elegans model also benefits from a strong community of researchers who have made nematode resources widely available. Recently, CeMbio was established, a set of twelve fully characterized bacteria from nine families that represent core microbiome of C. elegans, this tool is helping to enable in-depth experiments in this field.
A common misconception is that C. elegans destroys all the bacteria that it consumes, and thus, any found in its gut is accidental and pathogenic. Actually, in the wild, many micro-organisms (including bacteria, fungi, and viruses) exist within the worm’s intestine [19]. In fact, Zhang et al. recently showed that there is natural genetic variation in wild-type C. elegans which shapes their microbiome assembly – regulated by immune, xenobiotic, and metabolic signaling pathways [18]. This further makes the case for a personalized approach to medicine – it is as if your genetics have a pathogenic influence on your microbiome.
“Studying host-microbe interactions in C. elegans can be used to reveal the underlying, or ‘hidden’ factors, in many debilitating and progressive diseases.”
C. elegans is a well-established model for studying the immune system, and is a powerful way to study host-pathogen interactions. It has been found that the p38 pathway, which is conserved between humans and C. elegans, plays a pivotal role in microbiota-initiated immune protection, however, it does not impact microbial composition [20-22].
C. elegans is also a popular model for investigating the effect of the microbiome on the nervous system, and neurodegenerative disorders, because the simplicity of the model allows for study of pathways that are difficult to track in larger, more complex organisms. For instance, enteric bacteria are thought to influence the pathogenesis of neurodegenerative diseases, however, this is extremely difficult to study in humans [23]. A C. elegans model showed that this bacteria did upset protein homeostasis, but that the molecule butyrate stopped the progression of the bacteria, and the associated misfolding of proteins. Thus, butyrate-producing microbes are intriguing for both preventative and treatment strategies in neurodegenerative disease. Furthermore, C. elegans research has identified the dopaminergic system as a target for treating autoimmune disorders, by finding that the inhibition of a dopamine receptor (DOP-4 in worms, akin to D1 in humans) activates the the p38/PMK-1 MAPK pathway which, in turn, enhances immunity [24]. Overall, C. elegans is not yet fully optimized for MGBA research, but numerous C. elegans models of neurodegenerative diseases exist (such as ALS, Alzheimer’s, frontotemporal dementia, and Huntington’s disease) and many of which have all been linked to dysbiosis [25].
As previously discussed, synbiotics (combinations of prebiotics, probiotics, and postbiotics) show substantial benefits for preventing disease and increasing health. However, the molecular mechanisms underlying these effects remain poorly understood [26]. C. elegans are starting to be used as a powerful model to better characterize these mechanisms and can be used to screen for therapeutics that improve overall health. It is particularly pressing to understand these mechanisms due to the proliferation of pre-, pro-, post-, and syn- biotic products currently on the market. One of the main benefits of these products appears to be a more robust immune system, which is something that can be seen in C. elegans. For instance, preconditioning with exposure to the probiotic Bacterium Lactobacillus acidophilus strain NCFM was showed an enhanced immune response [27,28] and treatment with Lactobacillus rhamnosus Lcr35 was found hinder toxic infection with Candida albicans [29].
For anti-aging properties, Lactobacillus rhamnosus CNCM I-3690, was found to protect worms by increasing their viability by 30% and lifespan by 20% [30]. Importantly, transcriptomic analysis revealed that the increase in the worm’s lifespan correlated with the expression of the DAF-16/insulin-like pathway – a pathway which is conserved in humans.
This overview of C. elegans microbiome research presents an exciting future, one that takes a personalized approach to medicine using Patient Avatars (C. elegans that have been inserted with a human’s variant using CRISPR).
Bottomline: the use of these alternative animal model systems will help researchers understand which microbiome compositions are pathogenic and which are therapeutic.
![]()
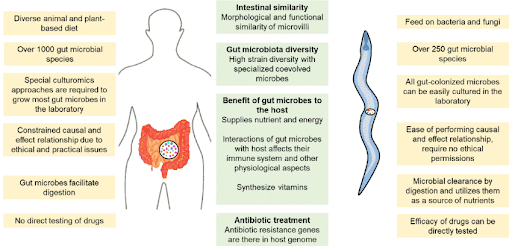
Figure 3. Differences (left and right sides) and similarities (center) between the functionalities of gut microbiota in C. elegans and humans [17].
Conclusion
The microbiome is a relatively new, but rapidly expanding, area of research. Formerly dominated by mammalian rodent studies, more and more researchers are finding that alternative models such as zebrafish and C. elegans can offer the ability to perform large-scale observational studies, with low maintenance cost, and yet not sacrifice translatability. As a result, modeling host-microbe interactions, dysbiosis, and disease-specific microbial communities in these organisms will help answer both fundamental questions about the role of microbes, and the underlying mechanisms involved in pathways, such as the cross talk between the microbes of the gut and response in the human brain, the MGBA axis. The microbiome may provide the link that is needed to understand autoimmune disorders such as coeliac disease, neurological diseases such as Alzheimer’s, and metabolic diseases including type 2 diabetes and non-alcoholic liver disease. This research is also intrinsically tied with examining the aging process, and as such, the gut microbiota could be the key to finding ways to improve human lifespans and healthspans.
To find out more details and learn more on how healthy microbiomes can be studied in animal models, please download our white paper at the link below.
In this white paper you will learn:
- The external effects (diet, environment, birth) on microbiome
- Microbiome products and therapies
- The functional foods and medicinal foods markets & the companies making them
- Regulatory concerns
- Traditional and alternative models to research microbiome
- More…
References
- Floor Hugenholtz WM de V. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci. 2018;75: 149.
- Zinöcker MK, Lindseth IA. The Western Diet-Microbiome-Host Interaction and Its Role in Metabolic Disease. Nutrients. 2018;10. doi:10.3390/nu10030365
- Craig H. Warden JSF. Comparisons of diets used in animal models of high fat feeding. Cell Metab. 2008;7: 277.
- Moysidou C-M, Owens RM. Advances in modelling the human microbiome-gut-brain axis in vitro. Biochem Soc Trans. 2021;49: 187-201.
- Wong S, Stephens WZ, Burns AR, Stagaman K, David LA, Bohannan BJ, et al. Ontogenetic Differences in Dietary Fat Influence Microbiota Assembly in the Zebrafish Gut. MBio. 2015;6. doi:10.1128/mBio.00687-15
- Stagaman K, Sharpton TJ, Guillemin K. Zebrafish microbiome studies make waves. Lab Anim . 2020;49: 201-207.
- Lickwar CR, Camp JG, Weiser M, Cocchiaro JL, Kingsley DM, Furey TS, et al. Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLoS Biol. 2017;15. doi:10.1371/journal.pbio.2002054
- Flores EM, Nguyen AT, Odem MA, Eisenhoffer GT, Krachler AM. The Zebrafish as a Model for Gastrointestinal Tract – Microbe Interactions. Cell Microbiol. 2020;22: e13152.
- Burns AR, Miller E, Agarwal M, Rolig AS, Milligan-Myhre K, Seredick S, et al. Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc Natl Acad Sci U S A. 2017;114. doi:10.1073/pnas.1702511114
- Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12. doi:10.1016/j.chom.2012.08.003
- Kim M, Benayoun BA. The microbiome: an emerging key player in aging and longevity. Transl Med Aging. 2020;4: 103-116.
- Stephens WZ, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K, et al. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 2016;10. doi:10.1038/ismej.2015.140
- Catron TR, Keely SP, Brinkman NE, Zurlinden TJ, Wood CE, Wright JR, et al. Host Developmental Toxicity of BPA and BPA Alternatives Is Inversely Related to Microbiota Disruption in Zebrafish. Toxicol Sci. 2019;167: 468-483.
- Gaulke CA, Martins ML, Watral VG, Humphreys IR, Spagnoli ST, Kent ML, et al. A longitudinal assessment of host-microbe-parasite interactions resolves the zebrafish gut microbiome’s link to Pseudocapillaria tomentosa infection and pathology. Microbiome. 2019;7. doi:10.1186/s40168-019-0622-9
- Lee J-G, Cho H-J, Jeong Y-M, Lee J-S. Genetic Approaches Using Zebrafish to Study the Microbiota-Gut-Brain Axis in Neurological Disorders. Cells. 2021 [cited 21 Oct 2021]. doi:10.3390/cells10030566
- Kulkarni S, Ganz J, Bayrer J, Becker L, Bogunovic M, Rao M. Advances in Enteric Neurobiology: The “Brain” in the Gut in Health and Disease. J Neurosci. 2018;38: 9346-9354.
- Kumar A, Baruah A, Tomioka M, Iino Y, Kalita MC, Khan M. Caenorhabditis elegans: a model to understand host-microbe interactions. Cell Mol Life Sci. 2020;77. doi:10.1007/s00018-019-03319-7
- Natural genetic variation drives microbiome selection in the Caenorhabditis elegans gut. Curr Biol. 2021;31: 2603-2618.e9.
- Hongbing Jiang DW. The Microbial Zoo in the C. elegans Intestine: Bacteria, Fungi and Viruses. Viruses. 2018;10. doi:10.3390/v10020085
- Association with Soil Bacteria Enhances p38-Dependent Infection Resistance in Caenorhabditis elegans. [cited 21 Oct 2021]. Available: https://journals.asm.org/doi/abs/10.1128/IAI.00653-12
- Ortiz A, Vega NM, Ratzke C, Gore J. Interspecies bacterial competition regulates community assembly in the C. elegans intestine. ISME J. 2021;15: 2131-2145.
- Host Immunity Alters Community Ecology and Stability of the Microbiome in a Caenorhabditis elegans Model. [cited 21 Oct 2021]. Available: https://journals.asm.org/doi/abs/10.1128/mSystems.00608-20
- Walker AC, Bhargava R, Vaziriyan-Sani AS, Pourciau C, Donahue ET, Dove AS, et al. Colonization of the Caenorhabditis elegans gut with human enteric bacterial pathogens leads to proteostasis disruption that is rescued by butyrate. PLoS Pathog. 2021;17: e1009510.
- Xiou Cao AA. Neural inhibition of dopaminergic signaling enhances immunity in a cell non-autonomous manner. Curr Biol. 2016;26: 2329.
- Lizett Ortiz de Ora ENB. Emergence of Caenorhabditis elegans as a Model Organism for Dissecting the Gut-Brain Axis. mSystems. 2021;6. doi:10.1128/mSystems.00755-21
- Poupet C, Chassard C, Nivoliez A, Bornes S. Caenorhabditis elegans, a Host to Investigate the Probiotic Properties of Beneficial Microorganisms. Front Nutr. 2020;0. doi:10.3389/fnut.2020.00135
- Kwon G, Lee J, Lim Y-H. Dairy Propionibacterium extends the mean lifespan of Caenorhabditis elegans via activation of the innate immune system. Sci Rep. 2016;6: 31713.
- Kim Y, Mylonakis E. Caenorhabditis elegans immune conditioning with the probiotic bacterium Lactobacillus acidophilus strain NCFM enhances gram-positive immune responses. Infect Immun. 2012;80: 2500-2508.
- Poupet C, Saraoui T, Veisseire P, Bonnet M, Dausset C, Gachinat M, et al. Lactobacillus rhamnosus Lcr35 as an effective treatment for preventing Candida albicans infection in the invertebrate model Caenorhabditis elegans: First mechanistic insights. PLoS One. 2019;14: e0216184.
- Grompone G, Martorell P, Llopis S, González N, Genovés S, Mulet AP, et al. Anti-Inflammatory Lactobacillus rhamnosus CNCM I-3690 Strain Protects against Oxidative Stress and Increases Lifespan in Caenorhabditis elegans. PLoS One. 2012;7: e52493.