As we all spend time isolating ourselves for social distancing purposes, group behavior is on the forefront of our minds in new and unforeseen ways. We as a species evolved to live and move in groups with complex social behaviors, and disruptions to these social interactions can impact us in profound and difficult ways, particularly with respect to our mental health. As we learn to navigate these changes in our day to day lives, perhaps now is a better time than any to collectively take some time to reflect on the ways mental health disorders impact some people every day, even when there isn’t a pandemic shutting down coffee shops and social gatherings.
Social processing deficits in psychiatric disorders like autism and schizophrenia are poorly understood, but researchers are taking steps to understand the underlying genetics and behaviors of these disorders. In a recent iScience publication, researchers at Harvard University and the Max Planck Institute of Animal Behavior used CRISPR-Cas9 to generate mutations in 90 different genes linked to human psychiatric disorders to investigate how they alter zebrafish group behaviors. After recording adult animals from each mutant line swimming in an open arena, videos were analyzed and variables like swimming speed, group spacing, and polarization were used to compare mutant fish against wild-type (WT) counterparts in a principal component analysis (PCA).
![]()
Figure from Tang et al., describing the principal components and impact on behaviors after accounting for speed (A) and illustrating the projection of each mutant onto the first 3 principal components, with sample projections for swimming behavior of each of the five highlighted mutants (B).
Authors found that differences in group swimming behaviors could largely be categorized into three patterns: scattered, huddled, and coordinated. Mutants in the scattered phenotype category included scn1lab and ctnnd2b – genes involved in epilepsy and autism – and were characterized by increased spacing between individuals and tendency toward dissociation and less collective coordination (1, 7). Mutants in the huddled phenotype category behaved with tighter spacing with more disordered behavior, clustering together as they traveled around the arena. Mutations in disc1 and immp2l – associated with schizophrenia and autism – had the most distinctive behavioral differences in this category (4, 2). Finally, mutants in the coordinated phenotype category were more likely to align their swimming direction and moved together more coherently, as the authors highlighted with a mutant in chrna2a, a gene implicated in epilepsy and addiction behaviors (6).
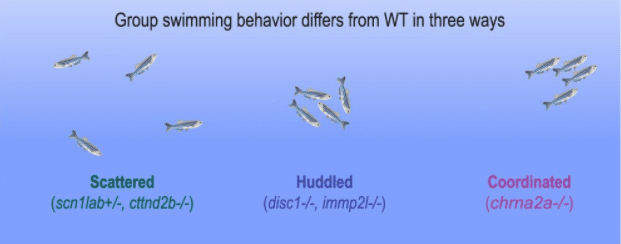
Figure from Tang et al., illustrating the distinct behavioral phenotypes identified and representative mutants in each category.
Authors noted that, for mutants that failed to produce a behavioral phenotype, they were unable to rule out the involvement of paralogous genes or compensatory gene expression induced in CRISPR-Cas9-generated mutations (5). Indeed, it would be worthwhile to see how mutants designed to avoid inducing such compensatory gene expression would compare in these assays (8). An additional limitation of this study was that while the effects of these mutations were generally described, the precise underlying mechanisms could not be elucidated, and will require higher resolution methods in the future to identify the morphological and cellular pathway changes induced by these mutations. Even with these limitations, though, this study is an impressive demonstration of the utility of zebrafish for interrogating the genetic basis of behavioral disorders, and will undoubtedly contribute to the discovery of both the underlying biology of human psychiatric disorders as well as novel pharmacological compounds for treating them.
Original Research:
- Tang, Wenlong, et al. “Genetic control of collective behavior in zebrafish.” iscience (2020): 100942.
References:
- Baraban, Scott C., Matthew T. Dinday, and Gabriela A. Hortopan. “Drug screening in Scn1a zebrafish mutant identifies clemizole as a potential Dravet syndrome treatment.” Nature communications 4.1 (2013): 1-10.
- Casey, Jillian P., et al. “A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder.” Human genetics 131.4 (2012): 565-579.
- Demontis, Ditte, et al. “Genome-wide association study implicates CHRNA2 in cannabis use disorder.” Nature neuroscience 22.7 (2019): 1066.
- Eachus, Helen, et al. “Disrupted-in-Schizophrenia-1 is essential for normal hypothalamic-pituitary-interrenal (HPI) axis function.” Human molecular genetics 26.11 (2017): 1992-2005.
- El-Brolosy, Mohamed A., et al. “Genetic compensation triggered by mutant mRNA degradation.” Nature 568.7751 (2019): 193-197.
- Hoda, Jean-Charles, et al. “Pleiotropic functional effects of the first epilepsy‐associated mutation in the human CHRNA2 gene.” FEBS letters 583.10 (2009): 1599-1604.
- Hofmeister, Wolfgang, et al. “Loss of zebrafish ctnnd2b results in disorganised forebrain neuron clusters.” bioRxiv (2018): 420828.
- Hoshijima, Kazuyuki, et al. “Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish.” Developmental cell 51.5 (2019): 645-657.



