You may or may not know that at InVivo Biosystems, we conduct a lot of grant funded research as well as offering custom models and assays to our clients. Much of our grant activity in the past few years has centered on creating and characterizing disease-associated variants and point mutations. We’ve created collaborations and partnerships with clinicians and researchers to identify genes and mutations that turn up again and again in diseases such as epilepsy, Alzheimers, and more.
Because CRISPR is the best gene editing method for creating small, precise edits, we use it to make all of our point mutations. We also use it to create “humanized animals”, which are genetically modified to have the human coding sequence for a particular gene, rather than the worm ortholog. Over the years we’ve been using CRISPR, we’ve refined our methods. We now use Co-CRISPR in almost all of our projects to make finding animals that have the precise edit easier.
As geneticists, we are naturally curious about how efficient CRISPR is. Recently, we took a step back to review the stats on point mutations we’ve created. Here’s a list of our big takeaways about how CRISPR gene editing is working for us:
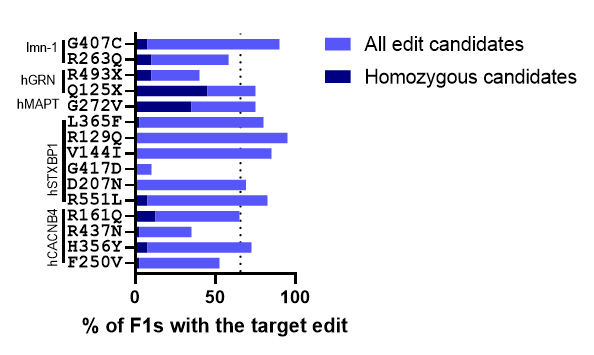
- CRISPR/Cas9 editing is typically very efficient. As we said above, we use Co-CRISPR to identify candidates in the F1 generation. Among the F1 candidates with the Co-CRISPR edit, our average percentage of animals with the target edit was 65.7%. Edit percentages as high as 95% were observed.
- Low co-CRISPR edit frequencies do not translate into low target edit frequencies. For two projects, only a few animals with the co-CRISPR phenotype could be identified for F1 screening (13 and 24 F1 animals, respectively. For reference, we usually screen 40 animals). However, for both of these projects the rate of target edits identified was high (69.2% and 58.3%). We might have thought that the low co-CRISPR edit frequency was related to the toxicity of the edit; however, it appears that was not the case for these edits. Don’t give up if your Co-CRISPR marker is rare!
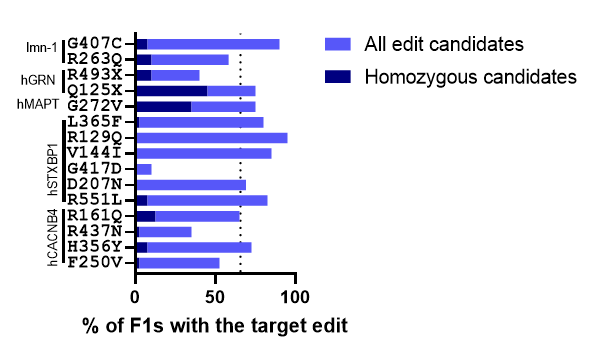
Figure 1. Target edit frequency in the F1 population. Animals with the co-CRISPR phenotype were isolated. After progeny production animals were harvested and lysed using C. elegans DNA extraction kit. Screening methods were used to identify animals containing the target edit. The percentage of animals containing the edit is indicated in light blue. The percentage of animals that were homozygous for the edit is indicated in dark blue. The dotted line indicates the average edit frequency of 65.7%. Data collected by Preston Kendrick and is also summarized in Table 1 below.
- Bi-alleleic conversion is observed. This was a big surprise: for 11 of the 15 projects, we were able to identify F1 animals that were homozygous for the target edit. On average, 9.5% of the F1 animals screened were homozygous for the target edit. This means that during the CRISPR injection, both alleles are being edited simultaneously. Which goes back to #1: CRISPR editing in our pipelines is very efficient.
- Each locus is unique. When we started this project, we might have thought that some genes would be easier to edit than others. It turns out that the site itself is more important than the gene. Even within the same gene, edit frequencies at different sites had a large range. For hCACNB4 the frequency of edits observed ranged from 35% to 72.5% at different loci within the gene. For hSTXBP1, the range was even bigger: One locus had an efficiency of 10% while another had 95% target edits.
- Point mutations can be generated quickly. For these projects, our average time from injection to sequence confirmation of two independent homozygous lines was 15.3 days. Our quickest project only took 10 days!
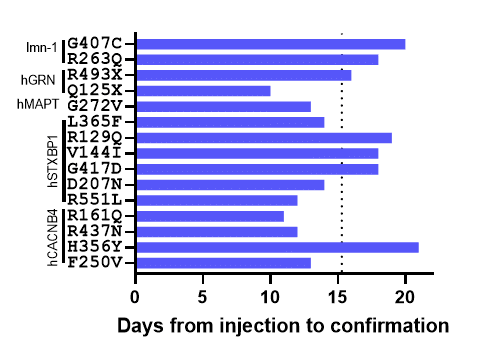
Figure 2. Time to generate homozygous sequence confirmed lines. The days from injection to confirmation is plotted for each project. The dotted line indicates the average time to completion of 15.3 days.
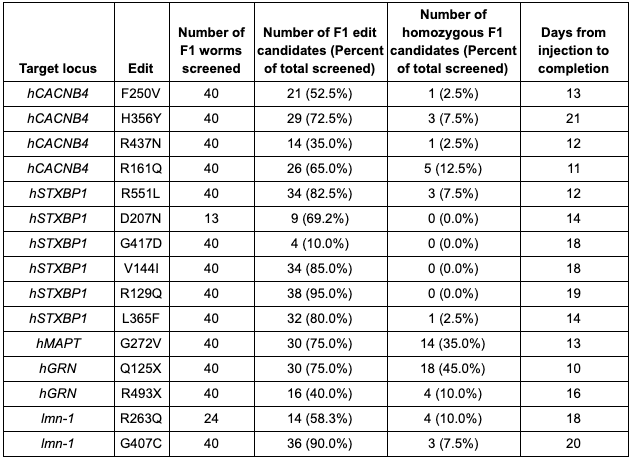
Table 1. Summary of data from Figure 1.
Learn more about our point mutation services and other C. elegans mutation service offerings