Fluorescent tagging of genes is widely used in many model organisms to study which tissues a gene of interest is expressed in, where in the cell a gene is expressed, or to determine whether multiple genes of interest are expressed in the same location. C. elegans is particularly well suited to fluorescence studies because they are transparent. Fluorescence can be easily observed in both live and fixed images.
At InVivo Biosystems, we have been working with fluorescent tags in C. elegans for over five years. In this span of time, we have made over 600 fluorescent transgenic lines. We have worked with 15 different fluorophores, including 8 variants of GFP.
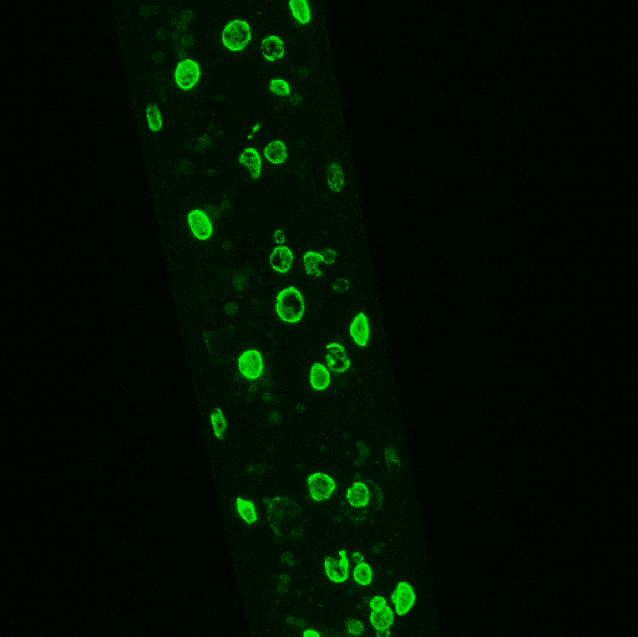
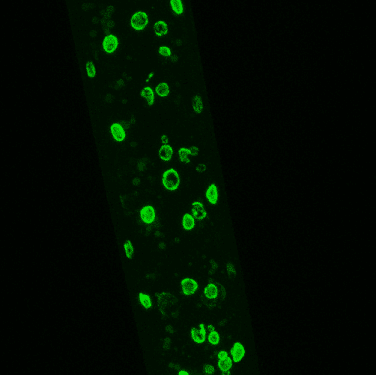
The image below is lmn-1 tagged endogenously with GFP showing a worm expressing our mCherry array markers, driven by the myo-2, myo-3 and rab-3 promoters.
Image 1. Fluorescent Tagging. IIntestinal nuclei from late L4 worms in an lmn-1::GFP transgenic strain. Image courtesy of Dr. André Catic, Baylor College of Medicine.
However, we are not limited to a fixed list of fluors. We are always happy to expand our repertoire of fluorescent tags, so if you have a fluor that you would like to use, please provide us the sequence and we will incorporate it into the design of your transgenic project. If you need your fluorescent transgenics lines to be imaged, we can readily help you with that too! We also have products, such as NemaGel, that may help you in imaging in your lab by immobilization of live animals. You can contact us if you have any questions.
In addition to fluorescent tagging, we also offer degron tagging and immunotagging services. Learn more about our C. elegans endogenous tagging services.