Summary:
Humanization of druggable targets is an important tool for increasing drug translation to clinic. Current in vivo drug discovery platforms can be quite limited in their ability to identify drug candidates which can successfully transition into humans because primary amino acid sequences are not conserved between animal models (mammalian mouse, zebrafish, and C. elegans), and humans [see Figure 1]. The differences between species’ amino acids means that there will be differences in the optimal drug binding site — this is where Whole-gene Humanized Animal Models (WHAM) can greatly improve drug discovery efficiency (patent US11477970B2; PMID 36411139).
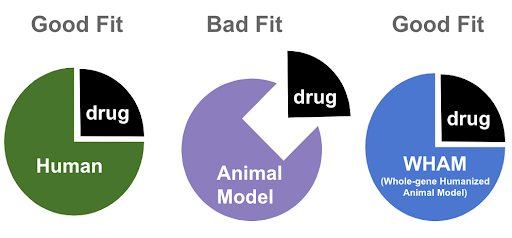
Figure 1. Binding Site Specificity: example of how designed molecules will have different binding site capacity between humans and animal models due to differences in primary amino acid sequence. When this amino acid difference is eliminated with WHAM humanization, better quality hits are identified from the animal model.
What is WHAM?
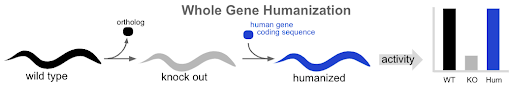
Figure 2. Whole-gene humanization technique. A native (WT) animal is CRISPR edited to make a gene KO animal and a whole-gene humanized animal has a gene swap replacement of ortholog locus.
Successful drug therapeutics are challenged for conserved activity between humans and animals: example from Cystic Fibrosis.
Currently, the most successful drug for treating Cystic Fibrosis (CF) was recently introduced to the market and it contains a 3-drug component mixture (Trikafta, made by Vertex Parmaceuticals). This mixture, which works by stabilizing the defects found in CF patients’ CFTR gene, has had a profoundly positive impact on CF patients (Hopkins et al., 2021; Laselva, Ardelean & Bear, 2021). Two components of Trikafta, made by Vertex Parmaceuticals, independently stabilize the CFTR chloride channel for transport to the plasma membrane (Elexacaftor and Tezacaftor), while the third component acts to modulate ion permeability leading to an increase residence time in open channel state (Ivacaftor) (Baatallah et al., 2021; Fiedorczuk & Chen, 2022). For one of the stabilizers, the binding site contacts of Tezacaftor are most optimal only with the human amino acids. When we look at animal model orthologs the binding site is not the same and the orthog’s inherent amino acid substitutions can create voids or steric hindrance which render the drug binding site of the ortholog gene as being suboptimal. For instance, in an extended analysis of the 22 amino acids involved in the Tezacaftor binding site, the conservation for the drug-contacting amino acids was found to be reduced in mice (77% identity), while both zebrafish and C. elegans were low in amino acid conservation (38% identity) (Baatallah et al., 2021). This indicates that the mouse gene would be a poor fit for Tezacaftor, and that the alternative fish and worm models contain enough variations that Tezacaftor may not bind at all.
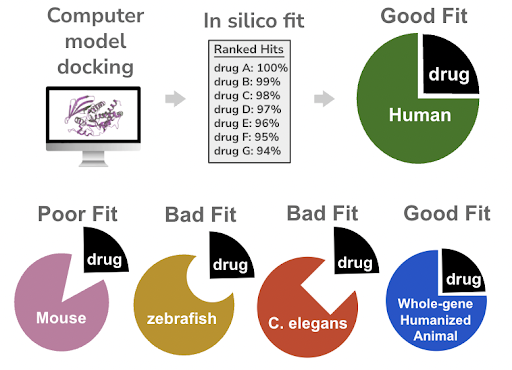
Figure 3. Discovery of drugs using computational approaches use human protein sequences, while screening of in silico hits in unmodified animal models can generate false negatives due to structural differences in drug binding site.
The lack of sequence conservation in drug binding sites is a shortcoming in drug screening on animal models. By analyzing what was successful for CF therapeutics in humans, we can observe how screening on animal-derived sequences has two major issues. The lack of sequence conservation between humans and animal models predicts that many of the hits found on an animal model will fail to work in humans (False Positives). Conversely, there would also be a set of compounds that were missed in the animal model but would have been found positive in humans (False Negatives). However, by using the WHAM method to bypass the molecular differences between species, the likelihood of achieving translation of hits-in-the-animal to winners-in-the-clinic will be much higher.
References:
Lopes-Pacheco M. CFTR Modulators: The Changing Face of Cystic Fibrosis in the Era of Precision Medicine. Front Pharmacol. 2019;10: 1662.
Capurro V, Tomati V, Sondo E, Renda M, Borrelli A, Pastorino C, et al. Partial Rescue of F508del-CFTR Stability and Trafficking Defects by Double Corrector Treatment. Int J Mol Sci. 2021;22. doi:10.3390/ijms22105262
Hopkins C, Onweni C, Zambito V, Fairweather D, McCormick K, Ebihara H, et al. Platforms for Personalized Polytherapeutics Discovery in COVID-19. J Mol Biol. 2021;433: 166945.
Laselva O, Ardelean MC, Bear CE. Phenotyping Rare CFTR Mutations Reveal Functional Expression Defects Restored by TRIKAFTA. J Pers Med. 2021;11. doi:10.3390/jpm11040301
Baatallah N, Elbahnsi A, Mornon J-P, Chevalier B, Pranke I, Servel N, et al. Pharmacological chaperones improve intra-domain stability and inter-domain assembly via distinct binding sites to rescue misfolded CFTR. Cell Mol Life Sci. 2021;78: 7813–7829
Fiedorczuk K, Chen J. Mechanism of CFTR correction by type I folding correctors. Cell. 2022;185: 158–168.e11.
Unlock the potential of drug discovery with our drug discovery and development services. Reach out to us now and expedite the development of life-saving treatments.




