Summary
The Undiagnosed Diseases Network UND is a nationwide network that is working to understand and diagnose rare and undiagnosed human diseases. The UND coordinates between research and clinical experts, using the latest genetic and genomic technologies to create a more personalized medicine model.
The Undiagnosed Diseases Network (UDN) was established in 2012 to bridge the divide between research and clinical care of unknown conditions (UDN, 2017). Rare diseases are actually quite numerous, affecting more than 25 million Americans and their families, putting a large strain on the healthcare system (NIH, 2020). It is estimated that 70-80% of undiagnosed diseases are due to rare genetic disorders, and so, the UND is working to understand mysterious conditions by pioneering a new personalized medicine model that uses genetic sequencing, model organism screening, and metabolics (UND, 2017).
Collaborative and Integrated work nature of the UDN
As its name implies, the UDN is a collaborative venture that brings together experts across the USA. The network is organized as three major sectors: the Coordinating Center, the Clinical Sites, and the Core Facilities (“Cores”) (Figure 1).
- The Coordinating Center: coordinates the overall UDN workflow (located at the Department of Biomedical Informatics at Harvard Medical School).
- The Clinical Sites: where UDN participants are further evaluated by a group of neurologists, immunologists, and geneticists to identify the root cause of the patient symptoms (located in 12 different cities across the United States).
- Core Facilities:
- The Sequencing Core: provides sequencing services for the UDN (Baylor College of Medicine).
- The Model Organisms Screening Center (MOSC): helps the network to understand how specific genetic changes contribute to disease by studying these changes in other organisms (Baylor College of Medicine, Washington University in St. Louis, and the University of Oregon).
- The Metabolomics Core: provides the UDN with advanced tools to study biological markers that might be related to disease (Mayo Clinic in Rochester, MN).
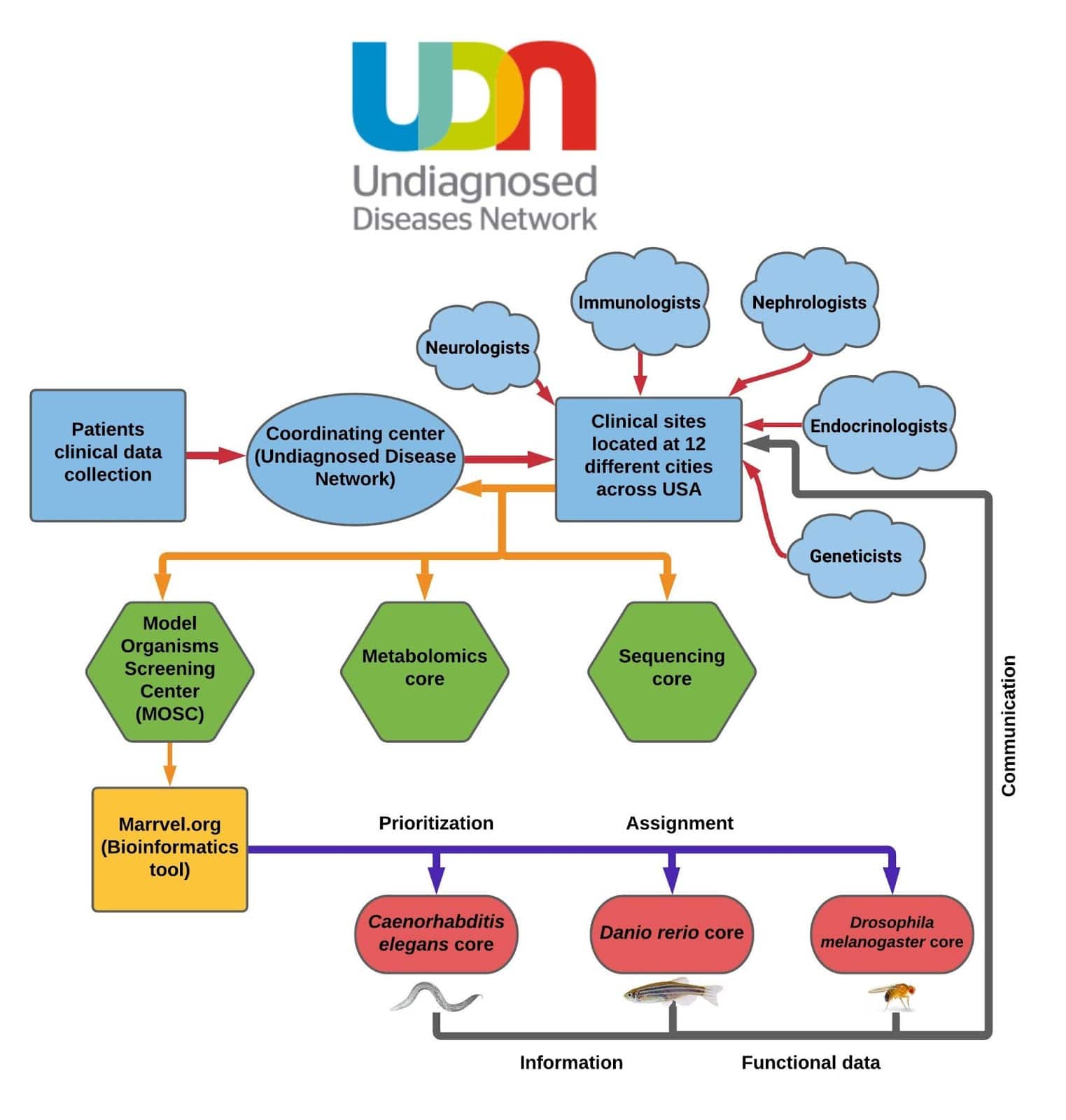
Figure 1: The UDN workflow chart depicting the following steps – 1) UDN clinical sites submit candidate gene(s)/variant(s) to the MOSCs – 2) MOSC utilizes the full potential of the bioinformatics tools, including the MARRVEL tool (marrvel.org) to aggregate the existing condition on the human gene/variant and its model organism orthologs. The MOSCs also matchmakes individuals with similar genotypes and phenotypes in other cohorts. 3) Once a variant is identified, MOSC scientists asses the gene assess gene and variant function of the target gene and further pursue in the C. elegans Core, Drosophila Core or Zebrafish Cores (https://nri.texaschildrens.org/udn).
The process of diagnosing an unknown condition begins when a patient applies to be a part of the UDN’s research study through the UDN Gateway. Patients undergo a thorough review process; then, those who are accepted (only 39.5% of applicants) either have their whole-exome, or whole-genome sequenced. In most cases, this analysis is also done on immediate family members. Sometimes, these comprehensive, genetic analyses provide the information needed to come to a diagnosis. However, if a diagnosis is not made, the Clinical Site will send the patient case to the The Model Organisms Screening Center (MOSC).
When the MOSC receives the patient case, their first step is to find any existing information on the gene/variant and its model organism orthologs using the MARRVEL database (Figure 2). The findings from the model organism research can then be rapidly translated into other studies such as investigations into higher order vertebrates, human tissues & cell culture models, and compound screening.
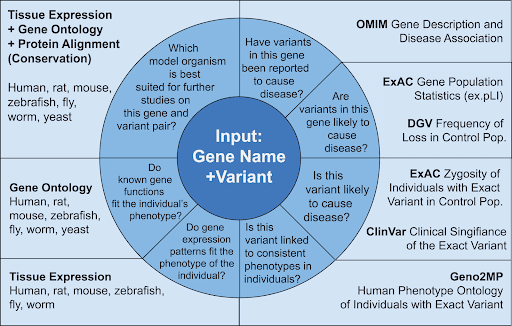
Figure 2: MARRVEL utilizes the unique approach of the gene variant analysis. Upon given commands by the user, the algorithm looks into the database starting at noon position of the inner ring, and then proceeds clockwise checking correct matches across different databases, producing the validated output to the end user (doi:10.1016/j.ajhg.2017.04.010).
Model organisms are essential to investigating the genetic basis of human diseases. Choice of model organism primarily depends on its evolutionarily conserved genes and biological mechanisms, which allow researchers to gain further insights into human biology. The UDN’s Model Organisms Screening Center uses three different model organisms: fruit flies (Drosophila melanogaster), nematode worms (Caenorhabditis elegans) and zebrafish (Danio rerio). These models were chosen because they are cost-efficient, have short life cycles, and scientists are capable of manipulating their genomes to translate the human disease condition.
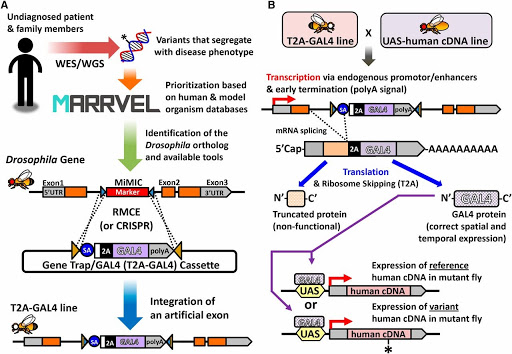
Figure 3: Strategy to “humanize” a Drosophila gene to assess functional consequences of a novel variant (https://doi.org/10.1534/genetics.117.203067).
The MOSC uses cutting-edge technology such as humanizing an orthologous gene in a model organism to determine whether a specific gene/variant is involved in the patient’s disease (Figure 3).
Thus far, the UDN has diagnosed 454 previously unknown diseases (UDN, 2021). Of these diagnoses, 81% were made through exome or genome sequencing, providing a strong case that the future of medicine is personalized, and reliant on a continuous dialogue between the clinical care and research fields.
In conclusion, the UDN serves a vital role in identifying the diagnostics of unknown disorders by collaborative work between multiple clinics, bioinformatics data analysis, MOSC, and the scientific community. The UDN is not the only network of its kind: Canada has a similar network called the Rare Diseases Models and Mechanisms network (RDMM), and other countries, such as India, uses GUaRDIAN network to study the genomics of rare diseases. There is also an international center to facilitate the rare disease network across the globe: Undiagnosed Diseases Network International (UDNI). By utilizing the fullest potential of gene editing studies in several model organisms, specifically C. elegans, Danio rerio, and Drosophila, we can translate results quickly to mammalian model systems and pluripotent stem cell research, leading to the identification of potential therapeutic targets and the specific genetic origin of the disease in humans.
References
- Children’s Hospital of Philadelphia (2021). About the Undiagnosed Diseases Network at CHOP. Children’s Hospital of Philadelphia Website. Retrieved from https://www.chop.edu/centers-programs/undiagnosed-diseases-network-chop/about
- Costa, Kevin (2019). The Importance of Water Quality in Keeping Zebrafish for Research. Blog: Hanna Instruments. Retrieved from https://blog.hannainst.com/water_quality_zebrafish
- Mullan, A. & Marsh, A. (2019). Advantages of using Drosophila Melanogaster as a Model Organism. Learning Centre: Oxford Instruments. Retrieved from https://andor.oxinst.com/learning/view/article/advantages-of-using-drosophila-melanogaster-as-a-model-organism
- NIH (2020). Undiagnosed Diseases Network: Highlights. National Institutes of Health, Common Fund Programs. Retrieved from https://commonfund.nih.gov/diseases/highlights
- Jan and Dan Duncan Neurological Research Institute (2021). The Undiagnosed Diseases Network (UDN). Texas Children’s Hospital. Retrieved from https://nri.texaschildrens.org/udn
- The Hardin Lab (2021). C. elegans. Retrieved from http://worms.zoology.wisc.edu/research/elegans/c_elegans.html
- Undiagnosed Diseases Network (2021). Quarterly Report: Winter 2021. https://undiagnosed.hms.harvard.edu/wp-content/uploads/2021/01/UDN-Quarterly-Report-Winter-2021.pdf
- Undiagnosed Diseases Network (2020). Undiagnosed Diseases Network (UDN): An NIH Common Fund Program. NIH: National Human Genome Research Institute. Retrieved from https://www.genome.gov/Funded-Programs-Projects/Undiagnosed-Diseases-Network
- Undiagnosed Diseases Network (2017). Model Organisms. Undiagnosed Diseases Network Website. Retrieved from https://undiagnosed.hms.harvard.edu/research/model-organisms-phase-ii/
- Wang, J., Al-Ouran, R., Hu, Y., Kim, S. Y., Wan, Y. W., Wangler, M. F., Yamamoto, S., Chao, H. T., Comjean, A., Mohr, S. E., UDN, Perrimon, N., Liu, Z., & Bellen, H. J. (2017). MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. American journal of human genetics, 100(6), 843-853. https://doi.org/10.1016/j.ajhg.2017.04.010
- Wangler, M.F., Yamamoto, S., Chao, HT. Posey, J.E., Westerfield, M., Postlethwait, J., and Members of the Undiagnosed Diseases Network (UDN) … (2017). Model Organisms Facilitate Rare Disease Diagnosis and Therapeutic Research, Genetics, 207(1)1, 9-27, https://doi.org/10.1534/genetics.117.203067



