Summary:
Over the past decade there has been an explosion in people experiencing gastrointestinal issues, and, consequently, there has been increased interest in research looking into the gut such as host-microbes interactions, the gut-brain axis, and our microbiome’s effect on our immune health. One of the conditions which has seen a rise in awareness is Leaky Gut Syndrome, however, there is disagreement among doctors about whether or not this condition actually exists. Many alternative practitioners say that Leaky Gut Syndrome is a real problem, while many Western doctors say that there is not enough evidence. Regardless of the condition’s validity, all medical practitioners agree that inflammation in your intestine (the basis of Leaky Gut Syndrome) is detrimental to your overall health, and can create long-reaching complications. So how is this phenomenon researched? Traditionally, mammalian mice models have been utilized, but there may be a better way. In this article we will discuss Leaky Gut, its causes and consequences, and the models being used to gain better insight into gut barrier disruption.
What is Leaky Gut?
Leaky gut is fairly self explanatory, literally it is an abnormal condition where leakiness starts to occur between your gut cells, more technically, it is defined as a gap in the intestinal junction barrier. Typically, your intestinal barrier is a semipermeable structure which enables your body to absorb essential nutrients while blocking against the entry of toxins such as pathogenic molecules and bacteria (Vancamelbeke & Vermeire, 2017). When there is loss of this critical barrier, it leads to higher permeability in the gut, where large molecules that normally would not be able to cross the gut barrier can now leak past and get into the bloodstream [Figure 1].
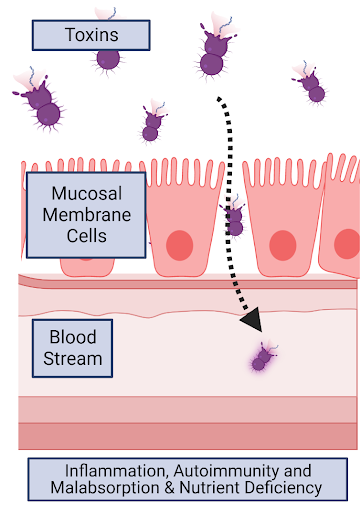
Figure 1. Leaky gut is defined as a gap in the intestinal junction barrier, this enables large molecules such as toxins to squeeze through the mucosal membrane cells and enter the bloodstream. This causes inflammatory responses which have large-reaching impacts on human health (made using BioRender).
What Causes Leaky Gut?
Leaky gut can be attributed to the consumption of material which irritates and inflames the gut lining tissue – this leads to inflammatory responses in gut tissues. As this inflammation accumulates and escalates, tissue necrosis starts and the once tight gap junctions between cells begin to form gaps,- the gut barrier begins to fail.
Many conditions can irritate the gut and trigger this inflammatory response. Common causes include bacterial and viral infections, chemotherapy and radiation which degrades the intestinal mucosa, chronic overuse of alcohol and Non-steroidal anti-inflammatory drugs (NSAIDs) such as aspirin and ibuprofen, as well as undesirable foods. These foods include any allergens an individual has, but also, more broadly, ultra-processed foods are well–known drivers of inflammatory responses, which is particularly concerning as ultra-processed foods have become the main source of calories in many countries: 58% in the USA, and nearly 50% in Canada and the UK (Hall et al., 2019; Heart & Stroke, 2017; Anthony, 2022).
What are the Consequences of a Leaky Gut?
Inflammation causes the loss of one’s nice, tight intestinal gap junction barrier which enables larger, and harmful materials to slip through the gut barrier; in turn, this leads to the disruption of various axes such as the gut-brain axis, gut-immune axis, and gut-cardio axis [Figure 2].
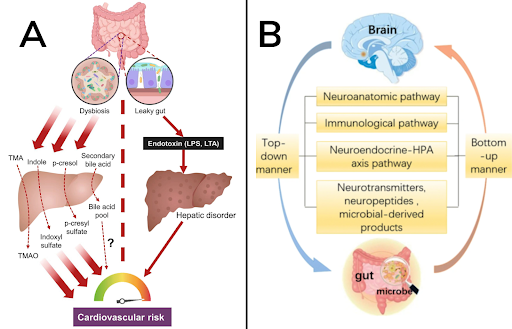
Figure 2. A) Gut-liver axis and cardiovascular risk. Dysbiosis leads to the influx of more gut-derived precursor metabolites, which are further metabolized by the liver to form cytotoxic chemicals to impair the cardiovascular function. In addition, leaky gut causes the leakage of endotoxin into the portal vein, damaging the liver function (Cheng & Huang, 2021). B) The gut-brain axis (GABA) a bidirectional link between the central nervous system and the enteric nervous system (ENS) of the body (Physiopedia, 2022).
Some studies have also suggested that malignant bacteria that enters the gut and travels to the liver can contribute to liver disease (Cleveland Clinic, 2022). Not only are these long-reach consequences, but the symptoms associated with Leaky Gut syndrome and the related inflammation, are highly detrimental to individuals’ life. Many people who suffer from this condition describe having brain fog, lethargy, bloating, cramping, burning and overall pain which makes them unable to perform daily tasks.
Leaky Gut Controversy?
Many doctors don’t recognize ‘leaky gut’ as a real condition. According to the majority of medical practitioners, Leaky Gut Syndrome is a ‘hypothetical condition’ that is merely based on the concept of intestinal permeability (Cleveland Clinic, 2022). When western practitioners do utilize the diagnosis, the term is often being used as a stopgap to cover anything related to Irritable Bowel Syndrome (IBS) (which, in of itself, is a catch-all for any discomfort associated with the gastrointestinal system that cannot be pinpointed). Despite the questions surrounding the validity of the pathological condition known as Leaky Gut Syndrome diagnosis, the gut permeability that is a consequence of tissue inflammation is a valid biomarker endpoint that can be quantified in Inflammatory Bowel Disease (IBD), which are chronic disease conditions localized to either the colon (ulcerative colitis) or at any location within the gut, but often at a localized hotspot (Crohn’s disease), where each can be distinguished by various blood biomarkers (Mitsialis et al., 2020).
“Leaky gut syndrome suggests that anything that injures your gut lining can lead to intestinal permeability (...) common everyday factors such as diet and stress may cumulatively wear down your intestinal lining until it becomes permeable. Scientists aren’t sure about this, but they do agree that these everyday factors may cause inflammation in your gut lining and uncomfortable GI symptoms for you.”
Cleveland Clinic, 2022
How Do You Measure Leaky Gut?
Like many aspects of human health, animals can be utilized as models of human structure. In this case, the gut barrier is largely conserved across species, both structurally and biochemically (González-González et al., 2019). Permeability to FITC-dextrans is commonly used in animal models to quantify changes in gut permeability. Exposure of the animal to gut irritant, then a pulse chase with a large fluorescently-labeled FITC-dextran molecule allows gut permeability to be quantified [Image 1].
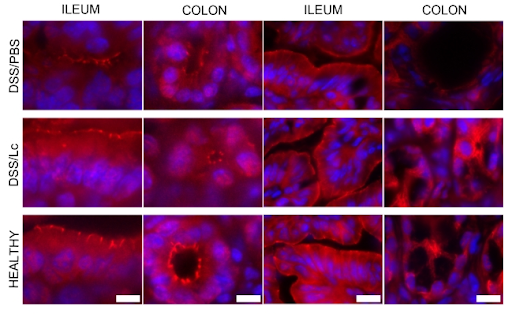
Image 1. Measurement of intestinal permeability by FITC-dextran – in order to determine whether probiotic could decrease severity of IBD in murine model (Zakostelska et al., 2011). Serum levels of 4.4-kDa FITC-dextran 5 hour after administration by gavage in DSS/PBS, DSS/Lc-treated group and healthy controls. Immunohistological detection of tight junction proteins ZO-1 (B) and occludin (C) in representative sections of colon and terminal ileum from DSS/PBS-, DSS/Lc-treated group and healthy controls. Fluorescent signal of ZO-1 or occludin (red) is merged with DAPI counterstained nuclei (blue).
What is the Most Common Model for Leaky Gut?
Like most biomedical fields, rodent models are typically used for research into gut barrier alterations, but since most animal models share the ability to display enhanced epithelial permeability, other models such as pig, horse, and birds models are commonly used as well.
Mus musculus (mice)
Mice models of intestinal injury range from studies showing burn injuries, exercise, and low-doses of radiation can all increase gut permeability (González-González et al., 2019). Many disease states manifest models with gut injury symptoms. For instance, in a mouse model of Huntington’s disease, loss of tight junction proteins was correlated with increased prevalence of leaky gut syndrome (Stan et al., 2020). Efforts to find biomarkers of leaky gut have led to the identification of calprotectin, a molecule released by neutrophils and macrophages that can be easily measured in fecal samples (Aranda et al., 2018). However, one of the most popular measures of leaky gut is the FITC-dextran assay method, as previously mentioned in Figure 1 (Zakostelska et al., 2011). Under the inflammatory conditions that can lead to leaky gut, the exposure of the intestinal lumen to FITC-dextran dye allows detection of leakage of dye into the gut tissues. Typically, rectal administration of dye is followed by blood draw three hours later for assessment of levels of FITC-dextran dye in blood by fluorescent spectrophotometer readings where high fluorescence reading in the blood quantify the severity of gut permeability to the dye (Cochran et al., 2020). Dextran sodium sulfate (DSS) is commonly used in animal models to induce leaky gut and the duration of exposure to 3% DSS is correlated with severity Cochran, Lamson & Whitehead, 2020).
Microbiota can have a significant impact on the formation of leaky gut. Lipopolysacarides (LPS) from bacterial origin are associated with both acute and chronic inflammation of intestinal tissues (Candelli et al., 2021). Infection often leads to elevated levels of C-reactive protein (CRP) and fecal calprotectin (FC), which are the two common biomarkers for IBD (Patel, Seif & Barrett, 2018). However more specific biomarkers are needed for detecting tight junction breakdown in the gut. One of the first biomarkers selected for this specificity was zonulin, a protein associated with triggering the breakdown in intestinal tight junctions (Fasano, 2012). However, its use as a biomarker has been hampered by the lack of specificity in current ELISA assays for the pre-HP2 allelic isoform (Massier et al., 2021). Further the amino sequence of the pre-HP2 only occurs in humans and therefore mouse studies on induction of intestinal permeability require transgenic mice expressing the pre-HP2 as a transgene. Galectins, OSM, and Non-coding RNAs are being explored for use as diagnostic markers (Chen et al., 2020).
Birds (Chicken)
Bird models, poultry in particular, are particularly useful in the study of intestinal permeability. For example, newly hatched chicks can be susceptible to invading pathogens that lead to inflammatory response in their gut. It was recently observed that parasites from Emeria species result in overexpression of ovotransferrin, one of the antimicrobial peptides in their gut and helps protect developing chicks from invading pathogens, and produces an inflammatory response (González-González et al., 2019). Similarly, local inflammation is increased in birds who are restricted feed for 24 hours (González-González et al., 2019).
Often, enteric inflammation models are created by orally administering a fluorescent probe (FITC-Dextran 4), and then observing different potentially inflammatory-inducing scenarios: for instance, being fed a high fat diet (HFD), Feed restriction (FRS), and Dextran sodium sulfate exposure (DSS) (Kuttappan et al., 2015).
Trickily, while DSS produces bird-inflammation models, when designing enteric experiments it is important to note that they are more sensitive to DSS than mammalian mouse models, and thus, there is a small bandwidth of proper dosage before DSS becomes toxic (Kuttappan et al., 2015).
Microphysiology systems (MPS) – ‘organ-on-chips’
A newer model, the advances in MPS over the past decade has produced fine tuning of cell culture techniques, fluid delivery systems, materials engineering, and performance enhancing modifications. As such, current MPS makes it possible to evaluate critical ADME parameters within a stand-alone organ system or through interconnected organ systems (Giordano, et al., 2021). Thus, MPS can be particularly useful in studying the interconnectivity of gut health, gut permeability, and the kidneys. The gut-kidney axis is well-known to have a significant impact on the immune system, and the development of chronic diseases [Figure 3].
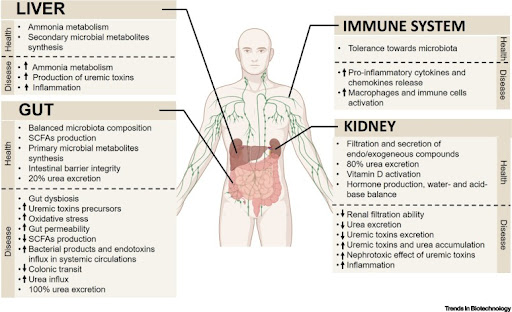
Figure 3. Gut–Kidney Axis Multiorgan Interactions in the Healthy State and in Chronic Kidney Disease (CKD) (Giordano, et al., 2021).
While it is still the case that animal models are the most common experimental models, cell culture systems are an attractive alternative, as it is extremely difficult to have highly controlled, and easily-visualized microenvironments in complex mammalian animal models.
What are Alternative Models for Leaky Gut?
While the previously mentioned mammalian models are traditionally used for research into the gut, there are limitations with the translatability of these models – as more and more it has become apparent the high impact that factors such as environment and diet has on the composition of microbiome, and overall gut health. Above, we mentioned MPS as a way to overcome some of the common hurdles that mammalian animal models introduce – such as their high complexity and low translatability. However, there are also inherent limitations in a non in vivo model.
One of the difficulties of investigating leaky gut is that the increased intestinal permeability produces a positive feedback loop: we can measure the increased inflammation and tightness of the intestinal gap, however, we are still unsure of what is the primary cause (Hamilton et al., 2022). This is where non-mammalian models such as the zebrafish and C. elegans worm offer exciting, game-changing alternatives. These models are notable as they provide in vivo ways to study the mechanisms behind leaky gut with many laboratory advantages, such as their short generation times, low cost, and high homology with humans.
zebrafish
Zebrafish have been an extremely successful model of gut dysbiosis, as they provide easy visualization of biological processes, and have conserved immune systems and intestinal function with humans. Since zebrafish have become an established model for microbial research their intestinal microbiota are already well-characterized, and there are readily available germ-free (GF) lines. This high level of control over the zebrafish gastrointestinal system enables researchers to reduce the level of inter-individuality between organisms which is often a limitation in microbiome research (Hamilton et al., 2022). In fact, zebrafish have already contributed to promising studies investigating intestinal barrier function. For instance, Marjoram et al. (2015), found that loss of function of epigenetic regulation increases intestinal epithelial cells and produces symptoms consistent with chronic inflammation. This finding indicates that mutations and dysfunction in epigenetic regulators can propagate the onset of symptoms which are characteristic of IBD (inflammatory bowel disease).
Although zebrafishes’ digestive system is functionally conserved, it should be noted that zebrafish do not have a stomach, but rather, have a digestive tract that is anatomically divided into separate sections: the mouth, the esophagus, three gut segments (anterior, middle, and posterior) [Image 4] (Wallace et al., 2005). And thus, zebrafish may be best posited as a high-value first model which can be used to cheaply and quickly investigate leaky gut before taking findings into a mammalian animal model.
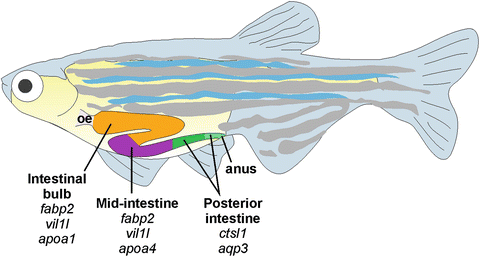
Figure 4. Anatomy of zebrafish stomach (Lobert, Mouradov & Heath, 2016).
Read more
C. elegans
Similar to the zebrafish, the small nematode, C. elegans, provides an in vivo model which enables a highly controllable, and well-defined microbe environment. Unlike zebrafish, however, C. elegans have a simple anatomy [Figure 5] – their gut is a simple 20-cell tube (Maduro, 2017). This makes C. elegans a good model to examine intestinal barrier function because, as previously noted, gut permeability is moderated by the intestinal epithelium, the single cell surface lining of both the small and large intestine, which is disrupted in leaky gut (Khong, Zhang & Zhang, 2018; Asano et al., 2003).
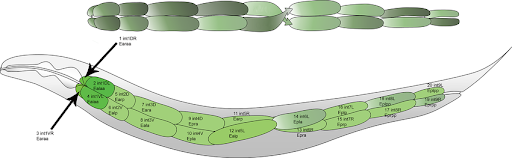
Figure 5. Layout of cells comprising the adult C. elegans intestine. (Mendenhall, 2015)
In fact, studies have found that successful therapeutic strategies for reducing intestinal inflammation in C. elegans translates into mice models (Wang et al., 2020). This indicates that despite their simplistic anatomy, therapeutics derived from studies using C. elegans can be applicable in more complex, mammalian models.
One of C. elegans’ greatest strengths, their simplicity, is also one of their biggest limitations, as they lack features of mammalian systems, such as a first-pass metabolism process in the liver, which can make translation more difficult. However, this being said, this limitation is able to be overcome by pairing C. elegans with more complex model systems.
Read More
Tight Junctions and C. elegans equivalents
The mammalian tight junction between epithelial cells of the intestine is a critical barrier to macromolecular entry into the gut. Similarly, C. elegans’ epithelial cells contain adhesion complexes [Figure 3]. However, C. elegans differ from mammals, vertebrates, and Drosophila in three unique ways: the worm cell-cell adhesion complex (the apical junction), has a single electron-dense area, LET-413 does not colocalize with DLG-1, and the tubular organs membranes’ include the proteins AR-3, PAR-6, PKC-3 and CRB-1. This being said, studies have found that C. elegans epithelial cells appear to abide by similar mechanisms as mammalian cells, making them suitable for modeling microbial dysbiosis, gastrointestinal inflammation, and intestinal disease pathogenesis (Sim & Hibbard, 2016; Böhm et al., 1997; Kim et al., 2022).
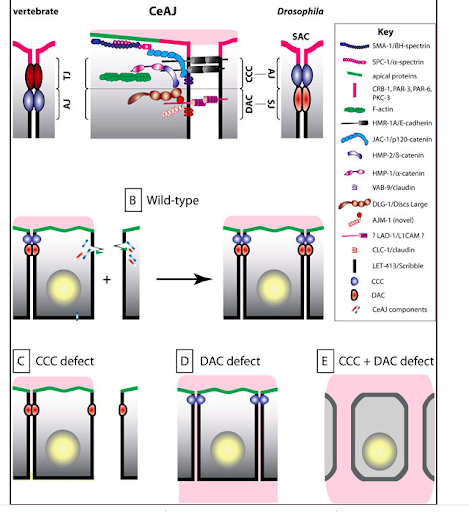
Figure 6. The CeAJ and cell-cell adhesion have analogous functions to vertebrate tight junctions (Labouesse, 2006).
Conclusion
The mammalian tight junction between epithelial cells of the intestine is a critical barrier to macromolecular entry into the gut. Similarly, C. elegans’ epithelial cells contain adhesion complexes [Figure 6]. However, C. elegans differ from mammals, vertebrates, and Drosophila in three unique ways: the worm cell-cell adhesion complex (the apical junction), has a single electron-dense area, LET-413 does not colocalize with DLG-1, and the tubular organs membranes’ include the proteins AR-3, PAR-6, PKC-3 and CRB-1. This being said, studies have found that C. elegans epithelial cells appear to abide by similar mechanisms as mammalian cells, making them suitable for modeling microbial dysbiosis, gastrointestinal inflammation, and intestinal disease pathogenesis (Sim & Hibbard, 2016; Böhm et al., 1997; Kim et al., 2022).
Leaky Gut, regardless of its validity as a medical diagnosis, is descriptive of real, life-impeding symptoms which are growing in prevalence. In order to better understand the mechanisms behind these symptoms and create therapeutics to alleviate these symptoms, animal models can be utilized. The empiric evidence obtained from these animal models will also have a larger-reaching impact on how we view the human body and its interconnected systems, as it has been shown that the gut influences nearly every aspect of human health.
As technology has improved, precision medicine has become possible, enabling tailored approaches to patients by utilizing an integrated multidisciplinary approach of genomic and environmental data. As leaky gut is involved in a multitude of pathologies, it is important to be able to address an individual’s specific symptoms.
Thus, this is an exciting and potentially monumental area of research. When designing a leaky gut assay, the choice of model is paramount. Alternative animal models, such as zebrafish and C. elegans offer fast, cost-effective to traditional mammalian models, which can accelerate breakthroughs in this area, and can promote better utilization of resources.
To learn more about InVivo Biosystems click here, or talk with one of our genome-editing experts today to see how we can aid in your research.
References:
- Vancamelbeke, M., & Vermeire, S. (2017). The intestinal barrier: a fundamental role in health and disease. Expert review of gastroenterology & hepatology, 11(9), 821–834. https://doi.org/10.1080/17474124.2017.1343143
- Cleveland Clinic (2022). Leaky Gut Syndrome, Cleveland Clinic, https://my.clevelandclinic.org/health/diseases/22724-leaky-gut-syndrome
- Zakostelska, Zuzana & Kverka, Miloslav & Kostovcikova, Klara & Rossmann, Pavel & Mrázek, Jakub & Kopecny, Jan & Hornova, Michaela & Srutkova,
- Dagmar & Hudcovic, Tomas & Ridl, Jakub & Tlaskalova-Hogenova, Helena. (2011). Lysate of Probiotic Lactobacillus casei DN-114 001 Ameliorates Colitis by Strengthening the Gut Barrier Function and Changing the Gut Microenvironment. PloS one. 6. e27961. 10.1371/journal.pone.0027961.
- Hall et al., 2019, Cell Metabolism 30, 67–77 July 2, 2019 Published by Elsevier Inc. https://doi.org/10.1016/j.cmet.2019.05.008
- Heart & Stroke (2017). News release: Time to curb our appetite for ultra-processed food, Heart & Stroke Canada, https://www.heartandstroke.ca/what-we-do/media-centre/news-releases/time-to-curb-our-appetite-for-ultra-processed-food
- Anthony, Andrew (2022). Fast food fever: how ultra-processed meals are unhealthier than you think, The Guardian, https://www.theguardian.com/science/2022/oct/16/ultra-processed-food-unhealthier-harder-to-avoid-than-you-thought
- Physiopedia (2022). Gut Brain Axis (GBA), Physiopedia, https://www.physio-pedia.com/Gut_Brain_Axis_%28GBA%29
- Cheng, C.K. & Huang, Y. (2021). The gut-cardiovascular connection: new era for cardiovascular therapy, Medical Review, vol. 1, no. 1, 2021, pp. 23-46. https://doi.org/10.1515/mr-2021-0002
- Labouesse, M. (2006). Epithelial junctions and attachments, WormBook, http://www.wormbook.org/chapters/www_epithelialjunctionsattach/epithelialjunctionsattach.html
- González-González, M., Díaz-Zepeda, C., Eyzaguirre-Velásquez, J., González-Arancibia, C., Bravo, J. A., & Julio-Pieper, M. (2019). Investigating Gut Permeability in Animal Models of Disease. Frontiers in physiology, 9, 1962. https://doi.org/10.3389/fphys.2018.01962
- Stan, T.L., Soylu-Kucharz, R., Burleigh, S. et al. Increased intestinal permeability and gut dysbiosis in the R6/2 mouse model of Huntington’s disease. Sci Rep 10, 18270 (2020). https://doi.org/10.1038/s41598-020-75229-9
- Kong, S., Zhang, Y. H., & Zhang, W. (2018). Regulation of Intestinal Epithelial Cells Properties and Functions by Amino Acids. BioMed research international, 2018, 2819154. https://doi.org/10.1155/2018/2819154
- Kuttappan VA, Vicuña EA, Latorre JD, Wolfenden AD, Téllez GI, Hargis BM and Bielke LR (2015) Evaluation of Gastrointestinal Leakage in Multiple Enteric Inflammation Models in Chickens. Front. Vet. Sci. 2:66. doi: 10.3389/fvets.2015.00066
- Giordano, L., Mihaila, S.M., Amirabadi, H.E., Masereeuw, R. (2021). Microphysiological Systems to Recapitulate the Gut–Kidney Axis., Trends in Biotechnology, Microphysiological systems, 39 (8) p811-823. https://doi.org/10.1016/j.tibtech.2020.12.001
- Marjoram, L., Alvers, A., Deerhake, M. E., Bagwell, J., Mankiewicz, J., Cocchiaro, J. L., Beerman, R. W., Willer, J., Sumigray, K. D., Katsanis, N., Tobin, D. M., Rawls, J. F., Goll, M. G., & Bagnat, M. (2015). Epigenetic control of intestinal barrier function and inflammation in zebrafish. Proceedings of the National Academy of Sciences of the United States of America, 112(9), 2770–2775. https://doi.org/10.1073/pnas.1424089112
- Hamilton, M. K., Wall, E. S., Robinson, C. D., Guillemin, K., & Eisen, J. S. (2022). Enteric nervous system modulation of luminal pH modifies the microbial environment to promote intestinal health. PLoS pathogens, 18(2), e1009989. https://doi.org/10.1371/journal.ppat.1009989
- Wallace, K. N., Akhter, S., Smith, E. M., Lorent, K., & Pack, M. (2005). Intestinal growth and differentiation in zebrafish. Mechanisms of development, 122(2), 157–173. https://doi.org/10.1016/j.mod.2004.10.009
- Lobert, Mouradov & Heath (2016). Focusing the Spotlight on the Zebrafish Intestine to Illuminate Mechanisms of Colorectal Cancer, Advances in Experimental Medicine and Biology book series (AEMB,volume 916), Cancer and zebrafish, https://link.springer.com/chapter/10.1007/978-3-319-30654-4_18
- Maduro M. F. (2017). Gut development in C. elegans. Seminars in cell & developmental biology, 66, 3–11. https://doi.org/10.1016/j.semcdb.2017.01.001
- Mendenhall, A. R., Tedesco, P. M., Sands, B., Johnson, T. E., & Brent, R. (2015). Single Cell Quantification of Reporter Gene Expression in Live Adult Caenorhabditis elegans Reveals Reproducible Cell-Specific Expression Patterns and Underlying Biological Variation. PloS one, 10(5), e0124289. https://doi.org/10.1371/journal.pone.0124289
- Aranda, C. J., Ocón, B., Arredondo-Amador, M., Suárez, M. D., Zarzuelo, A., Chazin, W. J., Martínez-Augustin, O., & Sánchez de Medina, F. (2018). Calprotectin protects against experimental colonic inflammation in mice. British journal of pharmacology, 175(19), 3797–3812. https://doi.org/10.1111/bph.14449
- Cochran, K. E., Lamson, N. G., & Whitehead, K. A. (2020). Expanding the utility of the dextran sulfate sodium (DSS) mouse model to induce a clinically relevant loss of intestinal barrier function. PeerJ, 8, e8681. https://doi.org/10.7717/peerj.8681
- Mitsialis, V., Wall, S., Liu, P., Ordovas-Montanes, J., Parmet, T., Vukovic, M., Spencer, D., Field, M., McCourt, C., Toothaker, J., Bousvaros, A., Boston Children’s Hospital Inflammatory Bowel Disease Center, Brigham and Women’s Hospital Crohn’s and Colitis Center, Shalek, A. K., Kean, L., Horwitz, B., Goldsmith, J., Tseng, G., Snapper, S. B., & Konnikova, L. (2020). Single-Cell Analyses of Colon and Blood Reveal Distinct Immune Cell Signatures of Ulcerative Colitis and Crohn’s Disease. Gastroenterology, 159(2), 591–608.e10. https://doi.org/10.1053/j.gastro.2020.04.074
- Patel, V., Seif, S., & Barrett, T. (2018). 2523 Noninvasive biomarkers for inflammatory bowel disease: Drawbacks and potential. Journal of Clinical and Translational Science, 2(Suppl 1), 22. https://doi.org/10.1017/cts.2018.102
- Candelli, M., Franza, L., Pignataro, G., Ojetti, V., Covino, M., Piccioni, A., Gasbarrini, A., & Franceschi, F. (2021). Interaction between Lipopolysaccharide and Gut Microbiota in Inflammatory Bowel Diseases. International journal of molecular sciences, 22(12), 6242. https://doi.org/10.3390/ijms22126242
- Massier, L., Chakaroun, R., Kovacs, P., & Heiker, J. T. (2021). Blurring the picture in leaky gut research: how shortcomings of zonulin as a biomarker mislead the field of intestinal permeability. Gut, 70(9), 1801–1802. https://doi.org/10.1136/gutjnl-2020-323026
- Chen, P., Zhou, G., Lin, J., Li, L., Zeng, Z., Chen, M., & Zhang, S. (2020). Serum Biomarkers for Inflammatory Bowel Disease. Frontiers in medicine, 7, 123. https://doi.org/10.3389/fmed.2020.00123
- Sim, S., Hibberd, M.L. Caenorhabditis elegans susceptibility to gut Enterococcus faecalis infection is associated with fat metabolism and epithelial junction integrity. BMC Microbiol 16, 6 (2016). https://doi.org/10.1186/s12866-016-0624-8
- Böhm, H., Brinkmann, V., Drab, M., Henske, A., & Kurzchalia, T. V. (1997). Mammalian homologues of C. elegans PAR-1 are asymmetrically localized in epithelial cells and may influence their polarity. Current biology : CB, 7(8), 603–606. https://doi.org/10.1016/s0960-9822(06)00260-0
- Kim, M. R., Cho, S. Y., Lee, H. J., Kim, J. Y., Nguyen, U. T. T., Ha, N. M., Choi, K. Y., Cha, K. H., Kim, J. H., Kim, W. K., & Kang, K. (2022). Schisandrin C improves leaky gut conditions in intestinal cell monolayer, organoid, and nematode models by increasing tight junction protein expression. Phytomedicine : international journal of phytotherapy and phytopharmacology, 103, 154209. https://doi.org/10.1016/j.phymed.2022.154209