Summary
Model organisms are essential to experimental research, allowing researchers and scientists to address a variety of questions. While no model is perfect, some models are more suitable for investigating specific questions than others. In this blog, we provide an overview of many commonly used model organisms, and compare their advantages and limitations.
Introduction
Throughout history animals have been used to model human anatomy and physiology. These model organisms are essential as they allow researchers to ask questions of biological processes, disease etiology and pathophysiology, and vaccine/therapy effectiveness (Beck & Meyerholz, 2020).
However, the choice of which model organism to use for a study should be carefully considered, as this can greatly impact the effectiveness of an experiment.
For instance, while research has largely utilized mammalian models such as rodents and apes, the use of these animals are often time-intensive, costly, and require close adherence to safety and ethics constraints.
The 3Rs of research, as described by W.M.S Russell and R.L. Burch (1959), have acted as a guiding principle for the humane treatment of animals for over fifty years. The 3Rs (Replacement, Reduction and Refinement) encourage researchers to consider alternatives to animals, use fewer animals, and minimize any distress they may encounter.
In recent years, advances in genome editing techniques have enabled a considerable step towards the replacement and reduction of animal research by expanding the available options of animal models to include Drosophila melanogaster (fruit fly), Caenorhabditis elegans (worm), and Danio rerio (zebrafish) (Beck & Meyerholz, 2020). These organisms provide an alternative to mammalian animal models, and can also be cheaper, faster, and more easily maintained.
Mammalian animal models still have a place in research, however, both from an ethical and logistical viewpoint researchers ought to consider all the model organisms available to them.
This article will address the strengths and limitations of many commonly used model organisms.
Cell Cultures
Cell culture is one of the most common laboratory tools used in life sciences, it consists of growing a cell (usually mammalian but can also be insect) in a controlled environment. Their frequent use has made cell cultures large contributors in groundbreaking scientific discoveries. For instance, research on the HeLa cell line alone has led to three Nobel Prizes (NIH, 2018). Significantly, in 2008, Dr Harald zur Hausen was awarded for his discovery of human papilloma viruses causing cervical cancer, which resulted in the development of a vaccine.
There are multiple types of cell culture: primary cell cultures (cells directly obtained from multicellular eukaryotes), secondary cultures (subcultures of primary cells; leads to the generation of cell lines), and cell lines (established cell culture) (Verma, Verma & Singh, 2020).
This variety in cell cultures has greatly advanced our understanding at a cellular level, allowing researchers to focus on a single cell type, studying in vitro in a highly controlled environment (Arango, et al, 2013).
While this model has greatly advanced research, it still has limitations such as requiring specific technical skills, and being susceptible to contamination. Additionally, there is often poor correlation between the findings (in vitro), and the biological mechanisms (in vivo).
Of course, the strengths and limitations of this model differ slightly between types of cell cultures: primary cell cultures are considered the best model for in vivo studies as they most directly correspond to the original eukaryote, having the same number and visual appearance of the chromosomes, but requiring some skill to obtain and having relatively short lifespans. Conversely, secondary cell cultures have much longer lifespans and are easy to cultivate, however, they can change overtime to become less representative of the in vivo state (Verma, Verma & Singh, 2020).
Drosophila melanogaster (fruit fly)
The Drosophila melanogaster has proven to be a very successful model organism, contributing to six Nobel Prizes with insights ranging from decision-making to circadian rhythms (Giamio, 2020). One of the reasons these insects have been so useful for research is their ease to breed and maintain, in fact, they are so easy to rear that they have been bred on a space shuttle to see how immune responses are altered by spaceflight (Marcu et al., 2011).
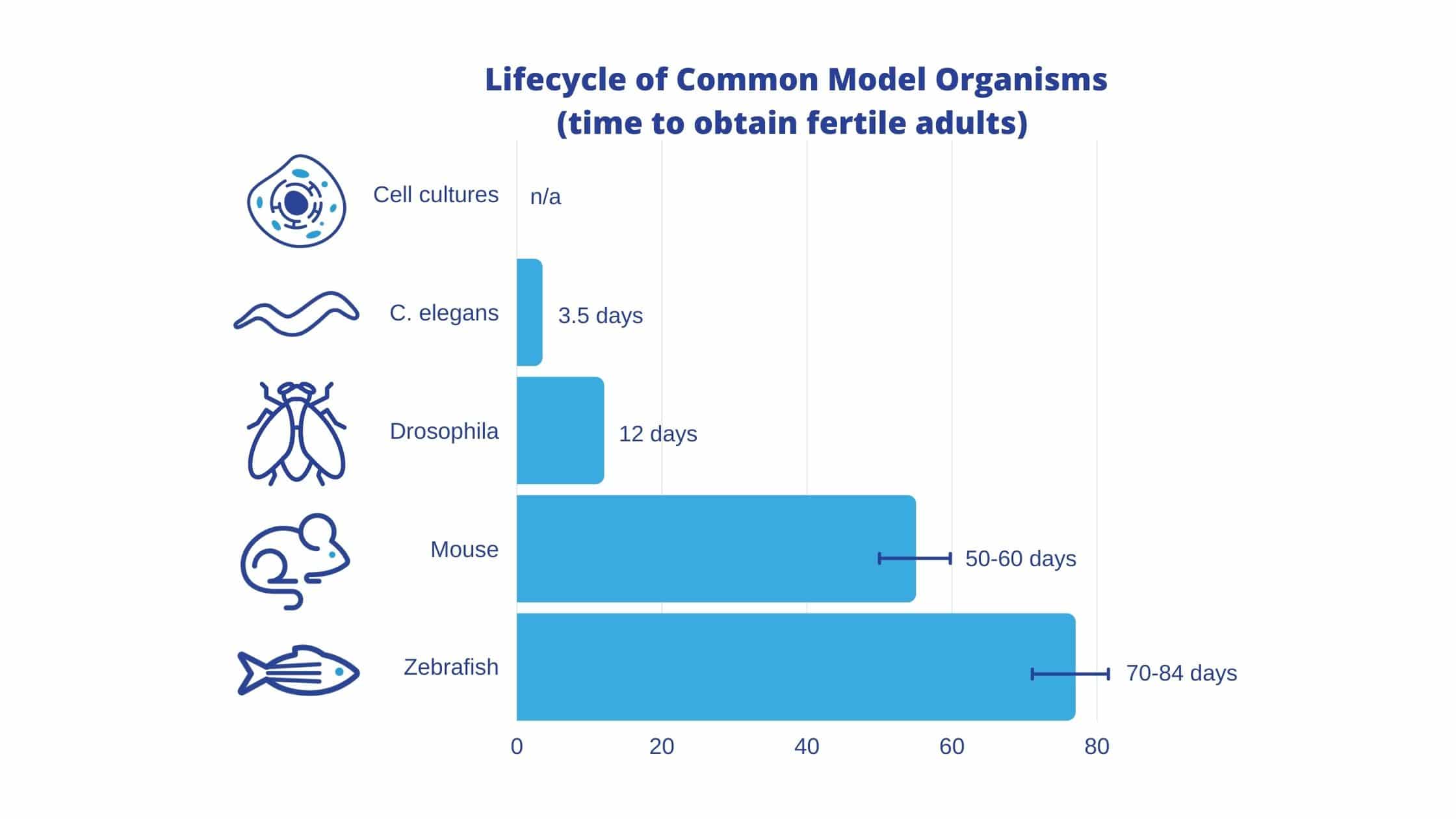
The fruit fly is easy to culture, and has a short lifespan (approx 12 days) (Nobel Media, 2020). The physical attributes of the fruit fly also make it a valuable model, as an adult fly’s organs and tissues act functionally equivalent to human organs and tissues (heart, lung, kidney, gut and reproductive tract) (Pandey & Nichols, 2011).
There are a few cell lines of the Drosophila melanogaster that are primarily used: Schneider lines, Kc16712, Mitsubishi/Miyake imaginal disc and central nervous system (CNS) lines, Milner laboratory imaginal disc lines, the adult ovarian cell lines, and the Ras lines (Luhur et al., 2019). As these lines depict, the fruit fly is a versatile model. Furthermore, there are many specific cell lines (over 160), and the fruit fly’s amendability to CRISPR-Cas9 genome editing means that it is possible to create new cell lines with specific genome modifications. The continued development of CRISPR technology has also led to new tsCRISPR (tissue-specific) tools which enable in vivo screenings (Meltzer et al., 2019).
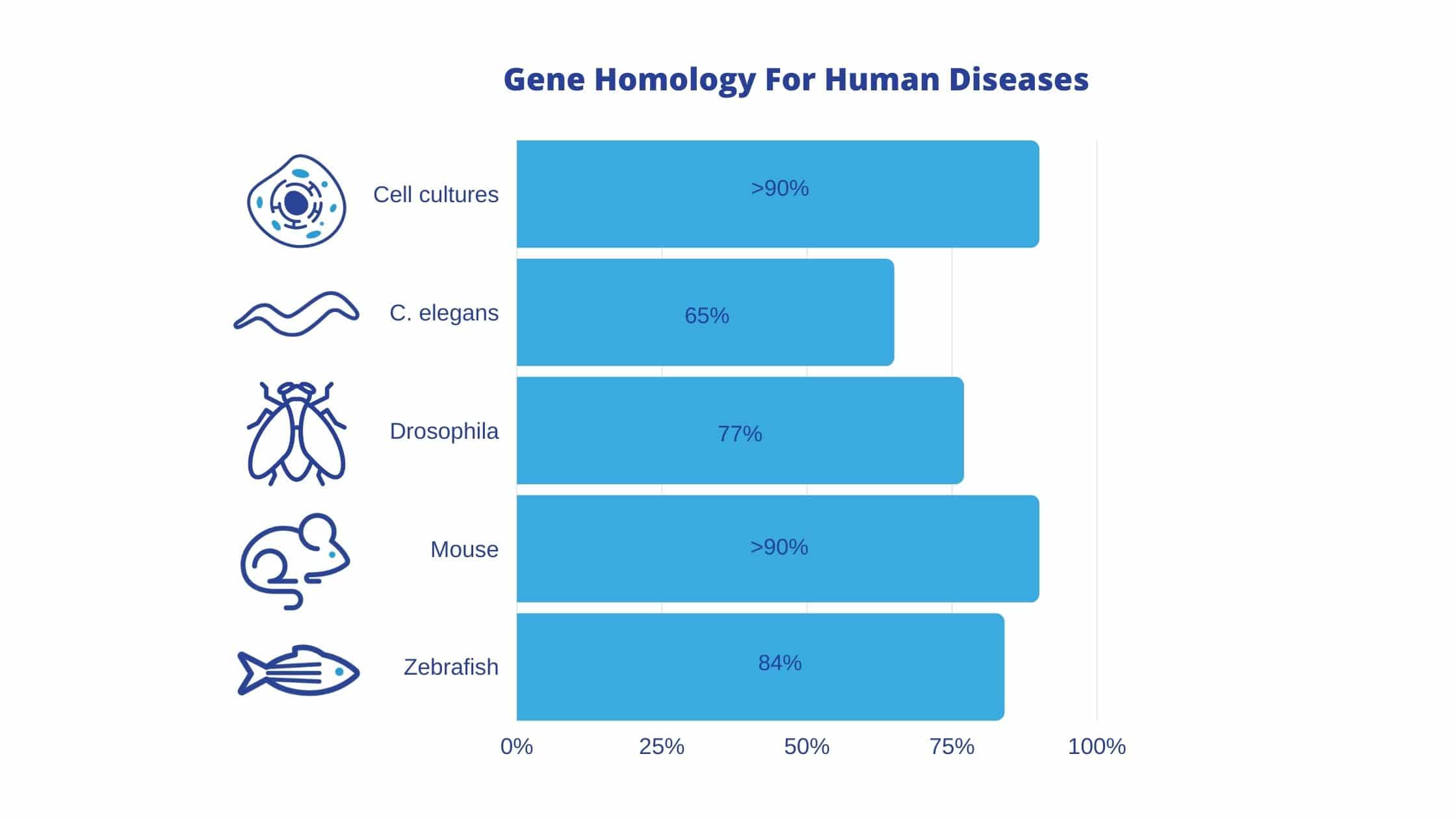
The fruit fly’s ability to be genetically manipulated so easily is especially useful because the Drosophila melanogaster has a counterpart for roughly 75% of genes associated with human genetic diseases (Ugur et al., 2016). Additionally, in 2000 the fly genome was fully sequenced and published, making it one of the most well understood model organisms.
A limitation of this model though is its inability to be frozen. Unlike mouse embryos or c. elegans, fruit flies have to be maintained as living stock, which requires researchers to frequently move them into new vials with food (Giamio, 2020).
In years to come, the Drosophila has the potential to make breakthroughs in our understanding of neurodegenerative diseases such as Huntington’s, Alzheimer’s and Parkinson’s, to further study the mechanisms behind conditions such as cancer and metabolic diseases, and to help develop targeted drug therapies.
Caenorhabditis elegans (worm)
Caenorhabditis elegans, more often referred to as C. elegans, are a more recently established model organism (1960s). However, six researchers have already won Nobel prizes based on their work using C. elegans: Sydney Brenner, John E. Sulston, and H. Robert Horvitz, (2002) for the discovery of genetic regulation or organ development and programmed cell death, Andrew Z. Fire, and Craig C. Mello (2006) for the discovery of RNA interference, and Martin Chalfie (2008) for the discovery of the green fluorescent protein, GFP (Van Der Linden Lab, 2020).
C. elegans have quickly been adopted in the research community thanks to their low cost, their ease to breed (>140 eggs per adult per day) and their ease to maintain (at 1mm, housing is extremely manageable) (Muschiol et al., 2009). In addition to their cost-effectiveness as live samples, C. elegans are able to be frozen and revived, which eliminates the cost and effort of maintaining colonies of mutants. Another beneficial property of C. elegans are their transparent bodies which allow for close observations of specific cells and overall development.
Like the fruit fly, C. elegans have a short life span (18-20 days), are easily genetically modified, and carry few ethical concerns (Zhang, et al., 2020). Also like the fruit fly, C. elegans have a completely sequenced genome; C. elegans was actually the first animal to be fully deciphered in 1998 (C. elegans Sequencing Consortium, 1998). While the C. elegans genome is only 40% homologous to humans, it is estimated that 65% of disease genes are homologous between the two species (Baumeister & Ge, 2002). This makes them a powerful tool for identifying human disease genes (Apfeld & Alper, 2018). In Particular, C. elegans are an ideal model for studying neurological diseases, as all of the neural populations in the human brain are represented in the C. elegans.
C. elegans have a simplistic body which has enabled researchers to fully identify all of its neurons (its connectome) which has only been accomplished in one other organism – the tadpole larvae of Ciona intestinalis (L.)(Ryan, Lu & Meinertzhagen, 2016). Notably, C. elegans can also be used for large-scale in vivo screenings, making it a valuable model organism in damage response (DDR) and DNA repair (DR) research (Honnen, 2017).
Another advantage of using C. elegans as a model organism is that there are already established protocols for using C. elegans in genetic toxicology and biomedicine. The “Million Mutations Project” published a library of 2007 mutagenized C. elegans strains in 2013 (Thompson et al., 2013). This unprecedented genetic resource identified mutant alleles for each of the worm’s 20,000 genes, with an average of 8 non-synonymous mutations per gene and more than 16,000 insertion/deletion and copy number changes (Thompson et al., 2013). C. elegans’ genes have been further annotated by the RNAi feeding library, which, in collaboration with Source BioScience, now targets approximately 87% of C. elegans genes (BioScience, 2020).
As for its limitations, one of C. elegans‘ strengths (its simplistic body) is also one of its greatest weaknesses. The C. elegans lacks some anatomical features of humans such as a brain, blood, defined fat cells, and internal organs, which prevents it from acting as a model for these mechanisms (Zhang, et al., 2020).
Thanks to the C. elegans‘ physiology, and our understanding of it, the C. elegans offers vast insights into the molecular mechanisms behind human diseases. C. elegans is able to be used in traditional genetic studies (forward genetics), as well as research that starts with the gene sequence (reverse genetics) (Corsi, Wightman & Chalfie, 2015). With developing technologies such as CRISPR, this model organism is surely going to lead to even more breakthroughs, as these gene editing techniques allow researchers to rapidly mutate and interrogate C. elegans genes.

Danio rerio (zebrafish)
The Danio rerio (formerly known as the Brachydanio rerio) is a freshwater fish typically found in the tropical waters of India, N. Pakistan, Nepal, Butan, and South Asia (Bozkurt, 2020). Dr Nüsslein-Volhardwas awarded the Nobel prize in 1995 for her part in the identification and classification of genes that control early embryonic development (Resnik & May, 2012). While Nüsslein-Volhard’s collaborators, Dr Lewis and Dr Wieschaus, mainly used fruit fly models, Nüsslein-Volhard extended the work to vertebrates. Finding zebrafish to be ideal for genetic and developmental studies she helped to make them a standard vertebrate research model. Since then, zebrafish’s prominence as a model organism has been on the rise: from 2008 to 2015 alone grants to fund zebrafish research increased almost 60% (Gaind, 2016). The zebrafish’s popularity is only expected to grow in future years as they are used in a wider range of studies.
The zebrafish’s small size (2.5-4cm), short life span (90 days maturation), and large brood size (200-300 eggs per week), make them sustainable, low maintenance, and contribute to their growing popularity as a model organism.
The transparency and ex utero development of zebrafish embryos are additional advantages of this model, allowing researchers to watch and manipulate the fish throughout development, unlike a mouse embryo which is opaque and contained within the mother mouse (NIH, 2016).
At this time, zebrafish are the only vertebrate that can be used for large-scale mutagenesis, and their use is well established: their genome is fully sequenced, and there are well developed methods for creating mutants using knock-in, knockdown, and knockout techniques (Teame et al., 2019). Zebrafish are an increasingly useful model, with new technologies, such as CRISPR-Cas9, being applied to speed up the transgenic process and make more mutant strains available (Burger et al., 2016).
Despite being a primitive vertebrate, zebrafish have all the main organs involved in metabolic processes, making them a valuable model for studying human disorders such as Type 2 diabetes mellitus, nonalcoholic fatty liver disease, and other hepatic diseases (Teame et al., 2019). Zebrafish also have the benefit of a similar macro-organization of the brain and cellular morphology to mammalian (mouse) models (Kalueff, Stewart & Gerlai, 2014). This similarity has made the zebrafish a popular model for studying complex brain disorders in humans, and has some exciting neuropharmacological prospects such as antiepileptic drugs, anxiolytic drugs, and SSRIs (selective serotonin reuptake inhibitors).
There are some limitations to the zebrafish as a model organism, however; as their lack of certain genes, and tissues/body parts, such as lungs, mammary glands, and prostate, mean that they are unable to act as models for diseases of these regions (NIH, 2016).
Zebrafish have already been successful models for a variety of human diseases such as Parkinson’s disease, Alzheimer’s, acute lymphoblastic leukemia, and human melanoma, among others (Teame et al., 2019). Thus, the zebrafish is a promising model for developing new preventative, and diagnostic care for human diseases, especially as new technologies allow us to create new mutant strains and circumvents some of this model’s previous limitations.
Mus musculus (Mouse)
Mouse models are considered the gold standard for research, and have contributed to the work of numerous Nobel prize recipients (Foundation for Biomedical Research, 2020). Most recently, William G. Kaelin Jr., Sir Peter J. Ratcliffe and Gregg L. Semenza were awarded for their discovery of how cells sense and adapt to oxygen availability (Nobel Media, 2019).
The recurrence of mice models in scientific breakthroughs is unsurprising, as the NADR (National Association for Biomedical Research) estimates that 95% of all warm-blooded animals used in laboratory research are rodents, and there is good reason for this (NADR, 2020). Rodents, and mice in particular, make extremely useful models: their genetic makeup is over 80% identical to humans, and they share many of the same brain functions as humans, which allows for behavioural research (Van Meer & Raber, 2005). Mice are also similar to humans in their spontaneous development of mutations, making them valuable models of heritable human diseases.
Since mice have a relatively short lifespan for mammals, they provide a way to model the progression of a disease from disease incubation and onset to the end-stage of the disease (NIH, 2020). That being said, there are exceptions depending on the mouse model: 15% of knockout mice (mice who have a ‘knocked out’ gene that researchers have replaced or disrupted) are unable to mature to full adulthood due to their genetically altered embryos (NIH, 2020). Also, in disease pathogenesis and drug therapy studies, these genetically engineered mice have the potential to display species-specific idiosyncrasies, making them unable to accurately work as a predictive model for human disease and treatment (Gurumurthy & Kent Lloyd, 2019).
Strategic failures in experimental design might be to blame for some of the mouse therapies that have failed to show results in humans, however, this points to one of the greatest limitations with a mouse model, and raises a larger conversation of laboratory mouse husbandry. When using a mouse model to test pathogens the temperature and microbial exposure of their environment should be considered. It has been suggested that the temperature the mice are being kept at is leading them to have cold stress, and their lack of environmental exposure to potential infections is unnatural (Karp, 2012). These are not insurmountable concerns, but should be addressed as both of these environmental factors may limit the translation value of these models. Mouse models also have significantly longer lifespans, and are significantly more costly than C. Elegans, fruit flies, and cell cultures.
Despite the previously noted concerns, mice are still extremely valuable as models for disease, as they, like humans, spontaneously develop mutations.
Conclusion
In deciding what model organism to use in a study, certain models are more adept for certain research objectives: for instance, the structural anatomy of zebrafish and C. elegans limit their ability to act as models for certain diseases, as they lack comparable structures to humans.
This being said, zebrafish have shown a remarkable similarity in their reaction to drugs as compared to rodent models. Currently, mice and rats are the most used for drug research, however, the comparable drug actions by rodents and zebrafish indicate that zebrafish could be an alternative model in neuropharmacology research.
Overall, the non-mammalian organisms discussed in this article (fruit flies, C. elegans, and zebrafish) are rapidly reproducing models with fully sequenced genomes and few ethical concerns. These non-mammalian organisms can act as valuable disease models, as the majority of genes known to be involved in human diseases have homologs in these organisms: 77% in fruit flies, 65% in C. elegans, and 84% in zebrafish. In comparison, disease genes in mammalian cell cultures and mice are 90% homologous to human disease genes.
Like the mammalian cell culture, the fruit fly, C. elegans, and zebrafish, are easily genetically modified. Thus, the continued advancement and refinement of new technologies, such as CRISPR, will make these models even more valuable for disease research and drug discovery.
Read Part 2: A Comparison of Common Organisms For Modeling Epilepsy
Bibliography
- Apfeld, J., & Alper, S. (2018). What Can We Learn About Human Disease from the Nematode C. elegans?. Methods in molecular biology (Clifton, N.J.), 1706, 53-75. https://doi.org/10.1007/978-1-4939-7471-9_4
- Arango MT, Quintero-Ronderos P, Castiblanco J, et al. (2013). Cell culture and cell analysis. In: Anaya JM, Shoenfeld Y, Rojas-Villarraga A, et al., editors. Autoimmunity: From Bench to Bedside [Internet]. Bogota (Colombia): El Rosario University Press; Chapter 45. Available from: https://www.ncbi.nlm.nih.gov/books/NBK459464/
- Baumeister, R., & Ge, L. (2002). The worm in us – Caenorhabditis elegans as a model of human disease. Trends in biotechnology, 20(4), 147-148. https://doi.org/10.1016/s0167-7799(01)01925-4
- Beck, A.P., Meyerholz, D.K.(2020). Evolving challenges to model human diseases for translational research. Cell Tissue Res 380, 305-311 https://doi.org/10.1007/s00441-019-03134-3
- Bozkurt, Y. (2020). Introductory chapter: Importance of ZEBRAFISH (DANIO RERIO) as model organism in biomedical research. Retrieved from https://www.intechopen.com/books/zebrafish-in-biomedical-research/introductory-chapter-importance-of-zebrafish-em-danio-rerio-em-as-model-organism-in-biomedical-resea
- Burger, A., H.Lindsay, A.Felker, C.Hess, C.Anders, E.Chiavacci, J.Zaugg, L.M.Weber, R.Catena, M.Jinek, et al. (2016). Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development, 143(11):2025-2037 doi:10.1242/dev.134809
- C. elegans Sequencing Consortium (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science (New York, N.Y.), 282(5396), 2012-2018. https://doi.org/10.1126/science.282.5396.2012
- C. elegans RNAi COLLECTION (AHRINGER): Source BioScience. (2020.). Retrieved from https://www.sourcebioscience.com/life-science-research/clones/rnai-resources/c-elegans-rnai-collection-ahringer/
- Corsi, A.K., Wightman, B., Chalfie, M. (2015). A Transparent Window into Biology: A Primer on Caenorhabditis elegans, Genetics, 200(2)1, 387-407, https://doi.org/10.1534/genetics.115.176099
- Foundation for Biomedical Research (2020).Nobel Prizes, Lab animals have made important contributions to nearly every Nobel Prize in Medicine. Retrieved from https://fbresearch.org/medical-advances/nobel-prizes/
- Gaind, Nisha. (2016). US grants for zebrafish studies on the rise. Springer Nature. Retrieved from https://www.nature.com/news/us-grants-for-zebrafish-studies-on-the-rise-1.20391
- Giaimo, C. (2020). Fruit flies are essential to science. so are the workers who keep them alive. Retrieved February 11, 2021, from https://www.nytimes.com/2020/12/14/science/fruit-flies-covid.html
- Gurumurthy, C.B & Kent Lloyd, K.C. (2019). Generating mouse models for biomedical research: technological advances. Disease Models & Mechanisms, 12(1), https://doi.org/10.1242/dmm.029462
- Honnen, S. Caenorhabditis elegans as a powerful alternative model organism to promote research in genetic toxicology and biomedicine. Arch Toxicol 91, 2029-2044 (2017). https://doi.org/10.1007/s00204-017-1944-7
- Introduction to cell culture. (n.d.). Retrieved from https://www.thermofisher.com/us/en/home/references/gibco-cell-culture-basics/introduction-to-cell-culture.html
- Kalueff AV, Stewart AM, Gerlai R. (2014). Zebrafish as an emerging model for studying complex brain disorders. Trends in Pharmacological Sciences;35(2):63-75
- Karp, C. L. 2012. Unstressing intemperate models: how cold stress undermines mouse modeling. J. Exp. Med. 209: 1069-1074.https://doi.org/10.1084/jem.20120988
- Luhur, A., Klueg, K. M., Roberts, J., & Zelhof, A. C. (2019). Thawing, Culturing, and Cryopreserving Drosophila Cell Lines. Journal of visualized experiments : JoVE, (146), 10.3791/59459. https://doi.org/10.3791/59459
- Marcu, O., Lera, M. P., Sanchez, M. E., Levic, E., Higgins, L. A., Shmygelska, A., Fahlen, T. F., Nichol, H., & Bhattacharya, S. (2011). Innate immune responses of Drosophila melanogaster are altered by spaceflight. PloS one, 6(1), e15361. https://doi.org/10.1371/journal.pone.0015361
- Meltzer, H., Marom, E., Alyagor, I. et al. (2019) Tissue-specific (ts)CRISPR as an efficient strategy for in vivo screening in Drosophila. Nat Commun 10, 2113. https://doi.org/10.1038/s41467-019-10140-0
- Meyerholz, D.K., Beck, A.P. & Singh, B. (2020). Innovative use of animal models to advance scientific research. Cell Tissue Res 380, 205-206 https://doi.org/10.1007/s00441-020-03210-z)
- NADR (2020) The importance of animal research. Retrieved from https://www.nabr.org/biomedical-research/importance-biomedical-research
- NIH (2020).Knockout Mice Fact Sheet. Retrieved from https://www.genome.gov/about-genomics/fact-sheets/Knockout-Mice-Fact-Sheet
- NIH (2018). Significant Research Advances Enabled by HeLa Cells. Retrieved from https://osp.od.nih.gov/scientific-sharing/hela-cells-timeline/
- Nobel Media. (2020). With a little help from our (insect) friends. Retrieved from https://www.nobelprize.org/drosophila/
- Nobel Media. (2019). Press release: The Nobel Prize in Physiology or Medicine 2019. Retrieved from https://www.nobelprize.org/prizes/medicine/2019/press-release/
- Pandey, U. B., & Nichols, C. D. (2011). Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacological reviews, 63(2), 411-436. https://doi.org/10.1124/pr.110.003293
- Resnik, Jack,, May, Catherine, “Christiane Nusslein-Volhard (1942- )”. Embryo Project Encyclopedia (2012-02-16). ISSN: 1940-5030 http://embryo.asu.edu/handle/10776/7901.
- Russell, W. M. S. and Burch, R. L. The Principles of Humane Experimental Technique. London: Methuen and Co. Ltd. (1959).
- Ryan K, Lu Z, Meinertzhagen IA. (2016). The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. Elife. doi: 10.7554/eLife.16962. PMID: 27921996; PMCID: PMC5140270.
- Thompson, O., Edgley, M., Strasbourger, P., Flibotte, S., Ewing, B., Adair, R., Au, V., Chaudhry, I., Fernando, L., Hutter, H., Kieffer, A., Lau, J., Lee, N., Miller, A., Raymant, G., Shen, B., Shendure, J., Taylor, J., Turner, E. H., Hillier, L. W., … Waterston, R. H. (2013). The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome research, 23(10), 1749-1762. https://doi.org/10.1101/gr.157651.113
- Van Der Linden Lab (2020). Why Worms. Retrieved from https://www.vanderlindenlab.com/why-worms
- Verma, A., Verma, M., & Singh, A. (2020). Animal tissue culture principles and applications. Animal Biotechnology, 269-293. https://doi.org/10.1016/B978-0-12-811710-1.00012-4
- Zhang, S., Li, F., Zhou, T., Wang, G., & Li, Z. (2020). Caenorhabditis elegans as a Useful Model for Studying Aging Mutations. Frontiers in endocrinology, 11, 554994. https://doi.org/10.3389/fendo.2020.554994