Summary
The development of CRISPR-Cas9 editing has allowed researchers to quickly create precise gene knockouts (KO), but while CRISPR-Cas9 KOs have been highly publicized, are the different methods understood? In this article we will discuss the two available methods for creating knockout models using the CRISPR-Cas9 technology: their advantages and limitations, and how they are being used in research.
In the study of biology, a system’s function can be better understood by disrupting it and observing the effects. This has become the go-to approach for many fields, from lesion studies in the brain, to knockout studies in genes. These knockout studies are a valuable component of the scientist’s toolbox – while many methods have been developed to manipulate genes, the advancement of technology has allowed gene disruption to become more precise. Notably, CRISPR-Cas9 enables researchers to target specific genes for removal at the genomic level. In this article we will discuss the two available methods of creating CRISPR-based knockout models- indels and full deletion- and the unique advantages and limitations of each, which should be taken into account when reading or conducting research.
Indels (one double-strand break)
The indel method gets its name from the INsertions and DELetions of genes in the genome. In this method, the Cas9 uses a single guide RNA (sgRNA) to target a region of the genome, and create a double-stranded break (Figure 1). The cell, sensing this double-strand break, goes about fixing the break by activating several DNA repair pathways. The indel method works because the cell’s repair process is not perfect, and often results in bases being inserted or deleted. When these insertions or deletions occur early in the coding sequence of the gene they can disable the gene entirely – as a frameshift leads to an early stop in the coding sequence.
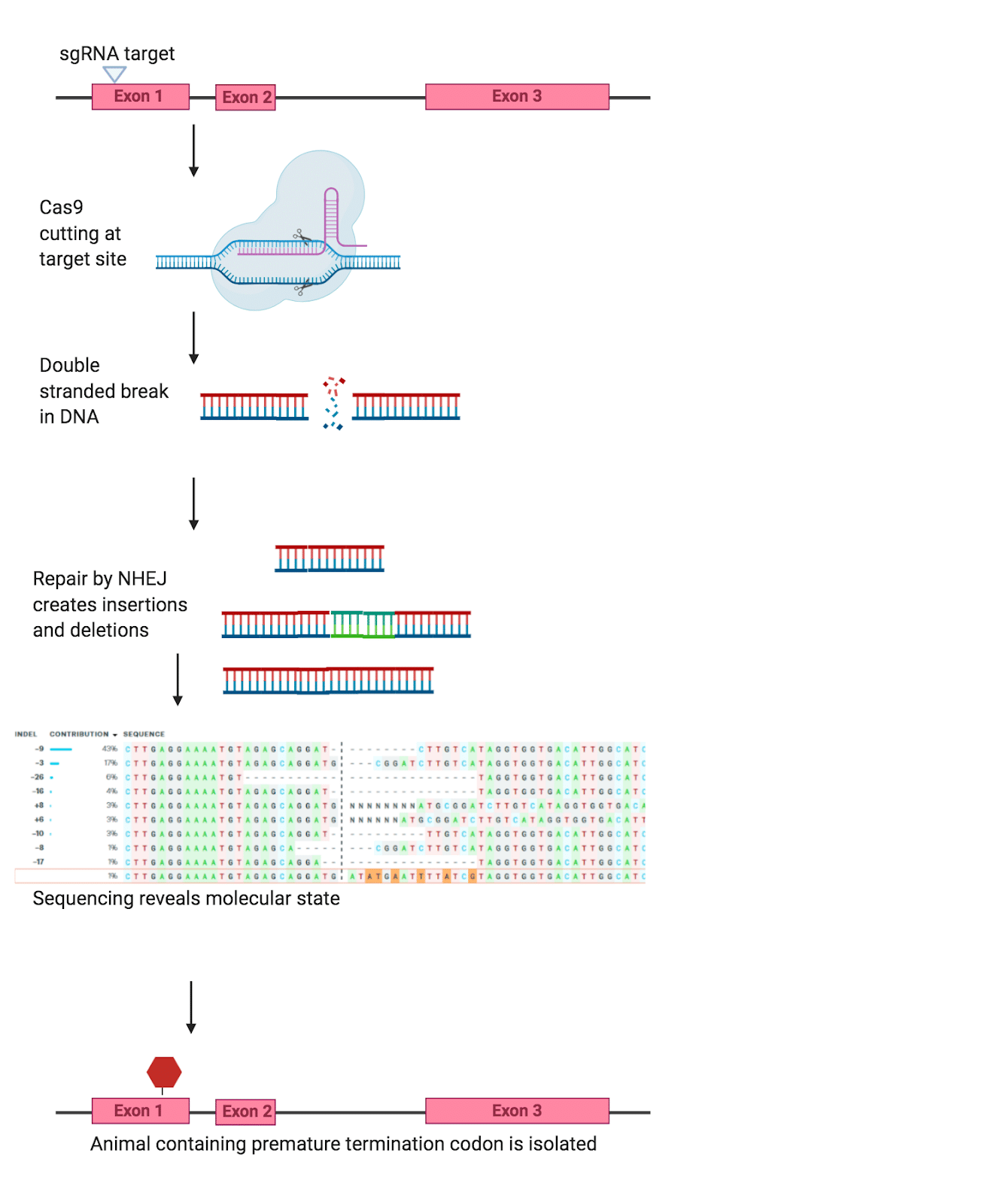
Figure One. Example of the Indel method (created in BioRender)
This method is highly effective for generating knockouts, in fact, you can even increase this method’s efficiency by using multiple sgRNAs all targeting the desired knockout. However, interpreting results from this method can be difficult as part of the coding sequence is still intact, with truncated proteins and alternative splicing having the potential to affect the gene. Furthermore, researchers have observed that the phenotypes seen with these models are not always as severe as expected due to transcriptional adaptation. Thus, despite how efficiently indel generation can create a knockout model, there are some drawbacks to this method.
Full deletion (two double-strand break)
In the full deletion method two sgRNA guides are used, one targeting each side of the to-be-deleted region of the genome. The Cas9 then creates two double-stranded breaks, this allows researchers to delete large regions of the coding sequence, or even the whole sequence When the cell goes about repairing these breaks it does so by joining together the remaining ends (Figure 2).
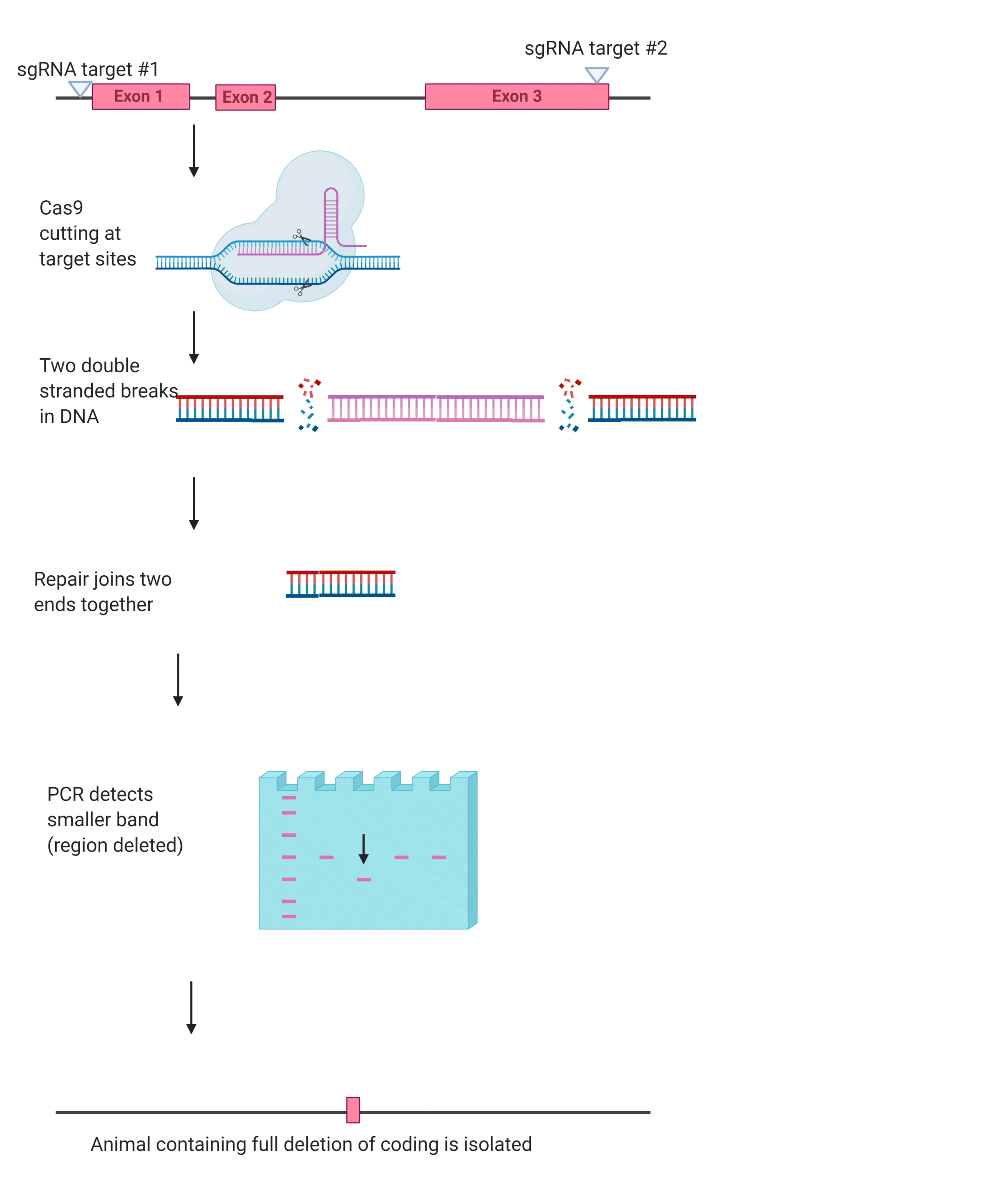
Figure Two. Example of the full deletion method (created in BioRender)
While it may seem like the additional sgRNA would make the full deletion method more effective, it is not as efficient as the indel method for creating knockouts due to the need to repair the breaks. This difficulty increases as the size of the to-be-deleted gene increases. However, sometimes the extra work that it takes to make a full deletion is worth it, as deleting the complete coding sequence means that there are no truncated proteins or splicing that can affect the gene’s subsequent loss-of-function.
Conclusion
Both of the methods described above to create a knockout model can result in the elimination of the desired gene. While both products work to create knockout models, careful controls need to be run to ensure researchers understand the consequences of the genetic manipulation; these may include protein quantification, mRNA expression analysis, and analysis of genetic compensation.
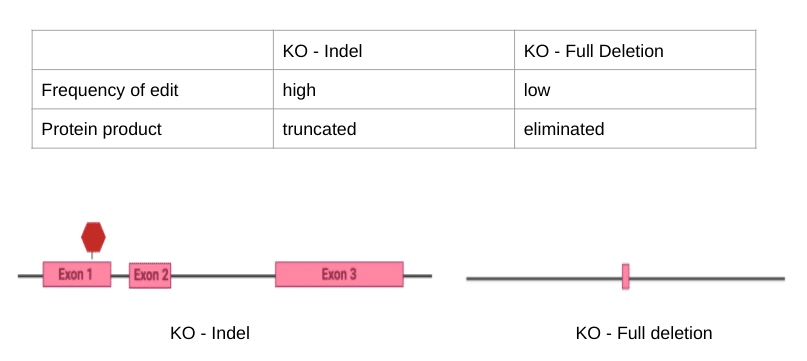
Figure Three. Comparison between the two types of KO techniques (created in BioRender)
Knockout models are valuable tools to better understand biology and disease, for instance, they enable researchers to create models of prevalent human disorders such as genetic disorders, which are often caused by a loss-of-function mutation in a gene. Additionally, reverse genetic approaches using knockouts have been invaluable to our understanding of biological processes. With more researchers utilizing CRISPR, it will be exciting to see all the new insights and understandings that come from our ever-expanding ability to manipulate the genome.
Examples
- Researchers made and characterized indel KO lines for 19 genes related to Fanconi anemia, a rare but serious blood disorder:
- Ramanagoudr-Bhojappa, R., Carrington, B., Ramaswami, M., Bishop, K., Robbins, G. M., Jones, M., Harper, U., Frederickson, S. C., Kimble, D. C., Sood, R., & Chandrasekharappa, S. C. (2018). Multiplexed CRISPR/Cas9-mediated knockout of 19 Fanconi anemia pathway genes in zebrafish revealed their roles in growth, sexual development and fertility. PLoS genetics, 14(12), e1007821. https://doi.org/10.1371/journal.pgen.1007821
- A Zebrafish consortium made indel KO strains for 636 Zebrafish genes on Chromosome 1:
- Sun, Y., Zhang, B., Luo, L., Shi, D. L., Wang, H., Cui, Z., Huang, H., Cao, Y., Shu, X., Zhang, W., Zhou, J., Li, Y., Du, J., Zhao, Q., Chen, J., Zhong, H., Zhong, T. P., Li, L., Xiong, J. W., Peng, J., … ZAKOC Consortium (2019). Systematic genome editing of the genes on zebrafish Chromosome 1 by CRISPR/Cas9. Genome research, 30(1), 118-126. Advance online publication. https://doi.org/10.1101/gr.248559.119
- These researchers made a 78kb deletion in Zebrafish to study smarca2, a gene involved in embryonic development and cancer:
- Kim, B. H., & Zhang, G. (2020). Generating Stable Knockout Zebrafish Lines by Deleting Large Chromosomal Fragments Using Multiple gRNAs. G3 (Bethesda, Md.), 10(3), 1029-1037. https://doi.org/10.1534/g3.119.401035




