Summary
Knockout and knock-in technologies enable researchers to modify genes in a chosen model system, and thus, can reveal a lot about how a gene functions. However, while these two methods may sound like complementary opposites, their purpose and design can actually have major differences. In this whitepaper we will discuss the available knockout and knock-in methods, and how they differ, so that you can determine which is best-suited for your experiment.
Knockouts vs. Knock-ins – what’s the difference?
As the name suggests, knockout (KO) models are generated by inactivating genes or gene segments, which typically leads to a loss of gene function. KOs frequently occur when random insertions or deletions (INDELs) are introduced into the genome by the inherently imprecise DNA repair process of non-homologous end joining (NHEJ) which is activated following double stranded breaks. Prior to CRISPR/Cas9, researchers relied on random mutagenesis to induce DNA changes following exposure to radiation or chemicals, such as ENU mutagenesis, which is a common method for forward screening in zebrafish (ingenious lab, 2018).
Knock-ins (KIs), in contrast, basically do the opposite. Rather than random disruption of already-present DNA sequences or genes, specific base pair changes are precisely introduced into the genome. Here, precise changes are directed by introduction of a template, e.g. a synthetic piece of DNA which is designed to replace an already existing genomic sequence, via the highly precise repair process of Homology-Directed Repair (HDR).
In addition to their functional differences, their purposes also differ. Knockouts are performed in order to observe downstream consequences, allowing that particular gene’s function, in addition to how it interacts with other genes, to be better characterized. Knock-ins typically introduce mutations that when present in humans are linked to genetic disease, allowing researchers to gain a better understanding of these mutations without human experimentation. Knock-ins can be used to introduce clinically relevant mutations linked to human diseases, or to test for effects of Single Nucleotide polymorphisms (SNPs) or variants of unknown function (VUSs) which could be benign or pathogenic.
Knockouts: Methods, Advantages and Limitations
There are two available methods of creating CRISPR-based knockout models: indels and full deletion. Both of these methods can result in the elimination of the desired gene. While both techniques work to create knockout models, careful controls need to be run to ensure researchers understand the consequences of the genetic manipulation; these may include protein quantification, mRNA expression analysis, and analysis of genetic compensation.
Indels (one double-strand break): The indel method gets its name from the INsertions and DELetions of genes in the genome. In this method, the Cas9 uses a single guide RNA (sgRNA) to target a region of the genome, and create a double-stranded break (Figure 1). The cell, sensing this double-strand break, goes about fixing the break by activating several DNA repair pathways. The indel method works because the cell’s repair process is not perfect, and often results in bases being inserted or deleted. When these insertions or deletions occur early in the coding sequence of the gene they can disable the gene entirely – as a frameshift leads to an early stop in the coding sequence.
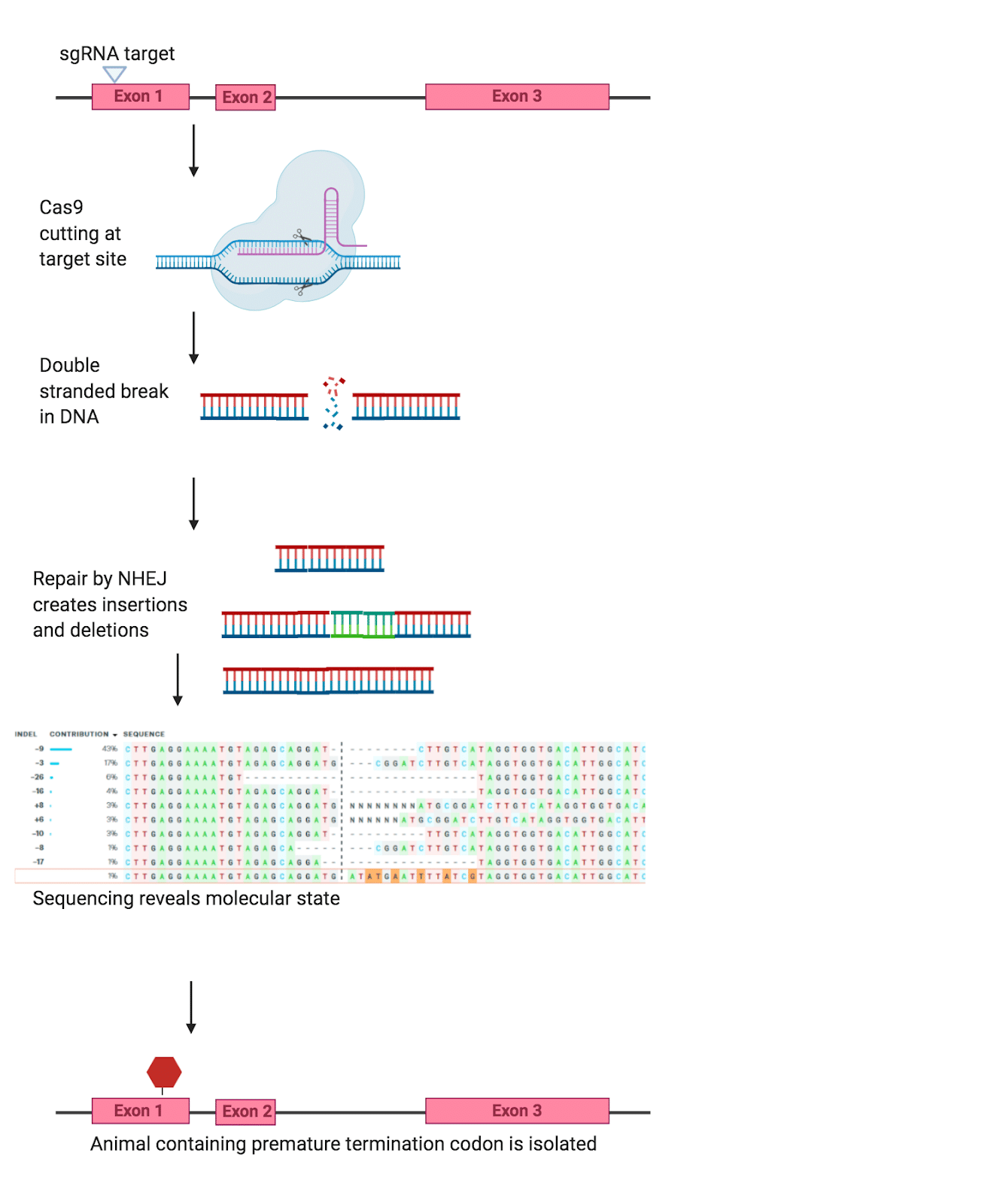
Figure One. Example of the Indel method (created in BioRender)
This method is highly effective for generating knockouts, in fact, you can even increase this method’s efficiency by using multiple sgRNAs all targeting the desired knockout. However, interpreting results from this method can be difficult as part of the coding sequence is still intact, with truncated proteins and alternative splicing having the potential to affect the gene. Furthermore, researchers have observed that the phenotypes seen with these models are not always as severe as expected due to transcriptional adaptation. Thus, despite how efficiently indel generation can create a knockout model, there are some drawbacks to this method.
Full deletion (two double-strand break): In the full deletion method two sgRNA guides are used, one targeting each side of the to-be-deleted region of the genome. The Cas9 then creates two double-stranded breaks, this allows researchers to delete large regions of the coding sequence, or even the whole sequence When the cell goes about repairing these breaks it does so by joining together the remaining ends (Figure 2).
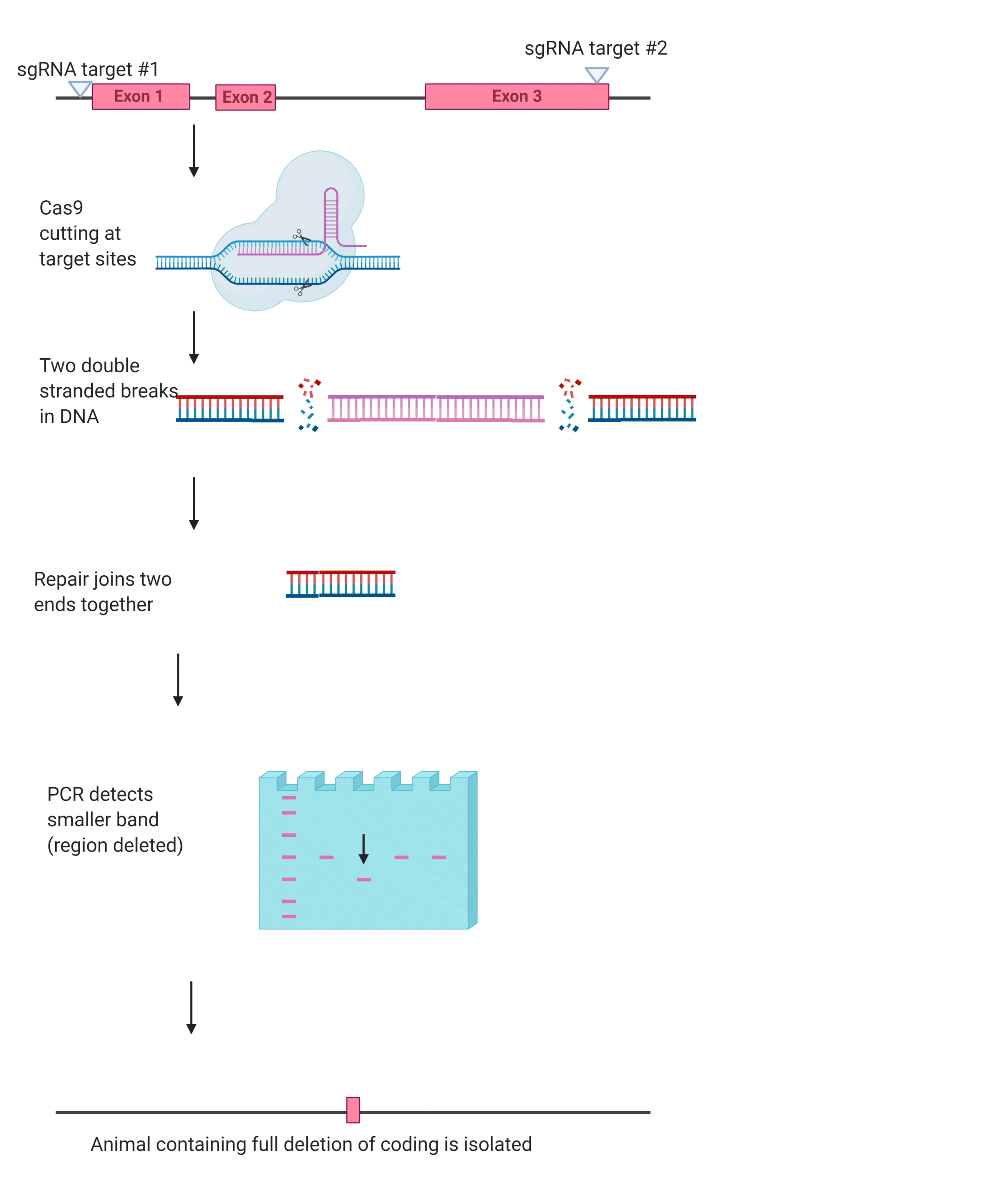
Figure Two. Example of the full deletion method (created in BioRender)
While it may seem like the additional sgRNA would make the full deletion method more effective, it is not as efficient as the indel method for creating knockouts due to the need to repair the breaks. This difficulty increases as the size of the to-be-deleted gene increases. However, sometimes the extra work that it takes to make a full deletion is worth it, as deleting the complete coding sequence means that there are no truncated proteins or splicing that can affect the gene’s subsequent loss-of-function.
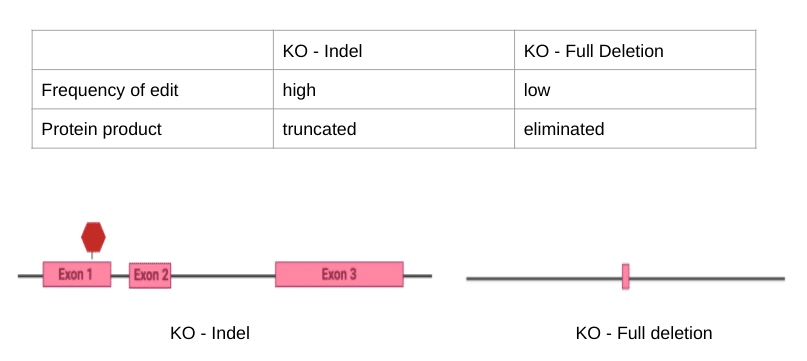
Figure Three. Comparison between the two types of knockout techniques (created in BioRender)
Knock-ins: Methods, Advantages and Limitations
The knock-in technology utilizes a CRISPR/CAS9 system to introduce a repair template containing the genetic sequence variants of interest. This method typically depends on the process of homology driven repair (HDR) to introduce a precise repair with an exogenously supplied sequence (Figure 4B. First, one or more single guide RNAs (sgRNA) direct Cas9 nuclease to make precise cuts in the genome at the location of the desired edit (Figure 4A). The sequence of interest, flanked by homology arms (sequences complementary to the regions surrounding the edit) then acts as a repair template, replacing the sequence that had been previously excised (Figure 4C). A tag may also be included in the inserted construct, which is a protein marker designed to allow reliable detection of the protein of interest.
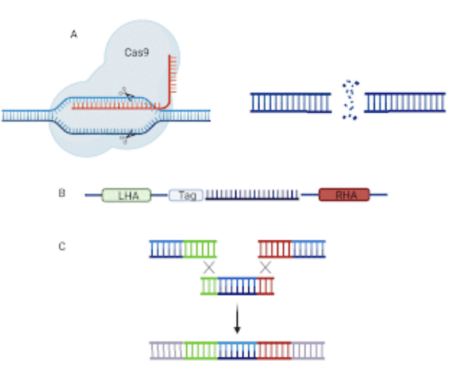
Figure Four. A: The sgRNA guides Cas9 to the proper binding site which then induces a DSB. B: The repair template is used to replace the excised DNA. C: The DSB is then repaired via HDR. (Made with BioRender).
There are many design considerations for using this system including both the type of edit to be introduced and the type of template to introduce that desired edit. When it comes to the type of edit, the design is pretty straightforward: researchers look towards the goals for the experiment. For example, epitope tags are often purpose built for detection of specific proteins through downstream applications, while fluorescent tags are commonly desired for following protein behavior in living cells or amino acid changes are used to test protein function or disease related single nucleotide polymorphisms (SNPs).
When it comes to the design of the template itself, things can get a little trickier. The specifics of the design can include which type of repair template to use (such as single or double stranded DNA or viral vectors), homology arm organization and specifications (length and symmetry around edit), and even the question of stranding (e.g. sense vs anti-sense) for single stranded templates. Here we will discuss some of these considerations and compare their respective advantages to determine which parameters may be best suited for your experiment.
Choosing the Right Template
Currently there are three types of repair templates receiving widespread use in knock-in editing including: double-stranded DNA (dsDNA) or Plasmid-based templates, short single-stranded DNA (ssDNA), and Adeno-associated viruses (AAVs) (Biswas & Enzmann 2021). Here, we’ll be focusing on dsDNA and ssDNA templates as AAV vectors are primarily used in gene therapeutics rather than in generating stable germline edited animals such as C.elegans or Zebrafish which is our specialty.
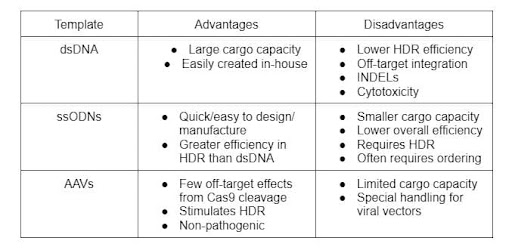
Table 1: Comparison of templates/vectors ( Biswas & Enzmann 2021, & Gaj et al. 2017)
Template choice is influenced by several factors with one being the size of the desired edit. For genome edits greater than several hundred base pairs (bp), dsDNA templates are primarily used (Table 1). At this size, production of ssDNA templates can be time-consuming with limited yields using current methodologies or prohibitively expensive and require ordering from dedicated manufacturers to ensure high quality, error-free templates (Iyer et al., 2019).
This increased size of dsDNA template translates to more options for the design of the desired insertion and perhaps a broader selection of tags or reporters. Two examples of dsDNA templates include plasmid-based templates and PCR product templates. For plasmid templates, the sequence of the desired edit is inserted into a plasmid backbone for ease of handling, propagation and microinjection. In addition to the increased cargo capacity of dsDNA templates, plasmid design and production can be done in most laboratories without specialty equipment or procedures that are currently demanded to produce long ssDNA templates (lssDNA) capable of the same cargo capacity (Table 1).
However, plasmid-based templates can result in random, off-target insertion of plasmid DNA into the host genome and have been found to possess higher rates of cytotoxicity than ssDNA template counterparts (Table 1). Despite the lower cargo capacity (on average <200bp), ssDNA templates (specifically single-strand oligodeoxynucleotides, or ssODNs) have demonstrated a greater efficiency in the HDR repair pathway in addition to lower cytotoxicity (Biswas & Enzmann 2021). Synthetic repair templates such as ssODNs have also demonstrated a lower rate of the off-target integration events found in the use of plasmid-based templates (Boel et al 2018).
Further Design Considerations
After selection of the template type, there are further design considerations to be made regarding overall template length, symmetry, and complementarity. The effects of these factors on HDR efficiency was examined by Boel et al. (2018) in which they targeted four sgRNA cut sites within four genes in zebrafish using ssODNs as repair templates. They found that increasing total template length from 60bp to 120bp across templates significantly improved the rate of HDR, while extending the template to 180bp generally resulted in a decrease of integration events. Template symmetry, that is differing lengths of left and right homology arms around the desired edit, and template complementarity to the target sequence were not found to have a significant impact on HDR rates. However it is unclear if this is generally true for all edited loci or specific to the one tested here.
Similar studies have been conducted and have given researchers a more comprehensive understanding of what variables to consider when designing a knock-in via the described CRISPR/CAS9 system, however template optimization is still hotly debated and limited systematic analysis has been completed, particularly in zebrafish. While there has been a large effort towards improving knock-in efficiency by tweaking knock-in template design, there remains little consensus on what is “best” and knock-in success rates remain frustratingly low, especially in zebrafish where it is common to see only >5% germline transmission rates.
Conclusion
With more researchers utilizing CRISPR, it will be exciting to see all the new insights and understandings that come from our ever-expanding ability to manipulate the genome. For instance, knockout models are valuable tools to better understand biology and disease – enabling researchers to create models of prevalent human disorders such as genetic disorders, which are often caused by a loss-of-function mutation in a gene. Additionally, reverse genetic approaches using knockouts have been invaluable to our understanding of biological processes. Furthermore, there is great promise in the advancing technology of knock-ins. Here at IVB we are exploring how template design can improve knock-in efficiency and have seen great success with introducing point mutations and small cargo such a loxP cassettes, especially in zebrafish where knock-in success can vary greatly from locus to locus. Given the large variety of genes we are targeting for our clients, we are also starting to identify some of the key design principles that maximize the chances for successful knock-in for our clients.
To talk to one of our experts about creating a custom knockout or knock-in model, contact us today.
Further reading:
- 11 Steps to a Successful CRISPR Knock-in
- What is Zebrafish Knock-in Tagging?
- The Challenge of Getting CRISPR-Based Knock-ins to Work in Zebrafish
- 6 Things That Can Go Wrong While Making Your Zebrafish CRISPR Knock-in
- The State of Knock-ins in Zebrafish
- How do Knock-ins Work?
- What’s a Knockout, Really?
- Precise Knockouts for Precise Research
- How Does Cre/LoxP Recombination Work?
- Differences Between HDR and NHEJ
Our CRISPR Knock-In Cell Lines service offers a precise and efficient way to introduce specific genetic modifications into your cells. With our cutting-edge technology and experienced team, we can create cell lines with the exact genetic changes you need for your research. Contact us today to learn more about our CRISPR Knock-In Cell Lines service and how we can help you achieve your research goals.
With the help of CRISPR knockout technology, you can modify genes precisely and efficiently, paving the way for groundbreaking discoveries in biotechnology. Download our whitepaper now and take the first step towards unlocking the full potential of knockout and knock-in technologies.
References:
- Ingenious lab (2018). Difference Between Knock In And Knockout, https://www.genetargeting.com/knockin/difference-between-knock-in-and-knockout/
- Biswas & Enzmann (2021). Optimizing CRISPR Knock-ins: Tips and Tricks For Successful Knock-in Editing, https://www.synthego.com/blog/crispr-knockin-tips-tricks
- Annekatrien Boel, Hanna De Saffel, Wouter Steyaert, Bert Callewaert, Anne De Paepe, Paul J. Coucke, Andy Willaert; CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis Model Mech 1 October 2018; 11 (10): dmm035352. doi: https://doi.org/10.1242/dmm.035352
- Gaj, T., Staahl, B.T., Rodrigues, G., Limsirichai, P., Ekman, F., Doudna, J.A., Schaffer, D.V., (2017). Targeted gene knock-in by homology-directed genome editing using Cas9 ribonucleoprotein and AAV donor delivery, Nucleic Acids Research, 45 (11), Pg. e98, https://doi.org/10.1093/nar/gkx154
- Iyer et al. (2019). Efficient Homology-directed Repair with Circular ssDNA Donors, http://dx.doi.org/10.1101/864199
About the Authors
Contributions to this piece were made by Karl Stutzner-Gibson, Trisha Brock, Marnie Preston, and Alexandra Narin.

