Expert CRISPR Knockout Service for Efficient Genome Editing
More C. elegans Models Services
InVivo Biosystems offers a comprehensive C. elegans CRISPR knockout gene editing service that includes the enhancement of genome editing using the CRISPR/Cas9 system.
Our customizable service allows for direct injection of the CRISPR/Cas9 system into mammalian and human cells, resulting in efficient and precise gene editing.
We also provide the recovery of custom genetic modifications, including knockouts and our service is competitively priced to make gene editing accessible to all. Our expertise in the CRISPR-Cas9 system also extends to bacterial immunity and the targeting of specific genomic loci for desired target effects.


In addition, our team has extensive experience in designing and implementing custom genetic modifications, including knock-ins, to suit your specific research needs.
We use Direct Injection to deliver the CRISPR-Cas9 system into mammalian and human cells, ensuring efficient editing and rapid recovery of custom cell lines. We can precisely insert genetic material at the target locus of your choice, allowing for targeted control of gene expression and study of target effects.
In addition, our team is skilled in working with embryonic stem cells, providing you with the opportunity to create transgenic models for a wide range of applications, from basic research to drug discovery.
Our C. elegans CRISPR Knockout services offer several advantages over traditional gene editing techniques, including:
- High efficiency: Precision genome editing enables the rapid generation of genetically modified cells and organisms.
- Specificity: Targeting specific genes and deactivating them with high accuracy, allowing for the creation of disease models and the discovery of gene function.
- Versatility: Our C. elegans genomes CRISPR Knockout services can be used in a wide range of applications, including drug discovery, gene therapy development, and genetic research.
- Speed and cost-effectiveness: Our CRISPR Knockout services are faster, more cost-effective, and require fewer resources than traditional gene editing techniques, allowing for more efficient and economical research.
Our C. elegans CRISPR Knockout services offer several advantages over traditional gene editing techniques, including:
- High efficiency: Precision genome editing enables the rapid generation of genetically modified cells and organisms.
- Specificity: Targeting specific genes and deactivating them with high accuracy, allowing for the creation of disease models and the discovery of gene function.
- Versatility: Our C. elegans genomes CRISPR Knockout services can be used in a wide range of applications, including drug discovery, gene therapy development, and genetic research.
- Speed and cost-effectiveness: Our CRISPR Knockout services are faster, more cost-effective, and require fewer resources than traditional gene editing techniques, allowing for more efficient and economical research.

Crispr Gene Knockout Process
Full, Partial And Conditional Gene Deletion
Full Gene Deletion
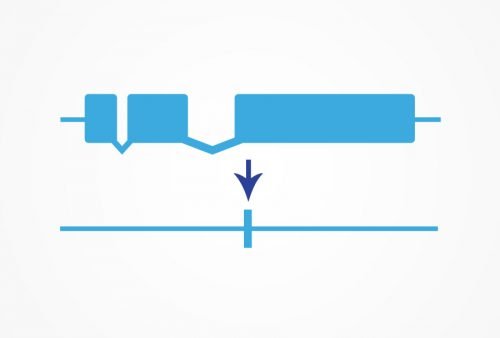
For research requiring a true null mutant, we recommend working with a precise KO to ensure the accuracy of your data. Many traditional null mutants rely on an early stop, which may not create a true null. Any genomic region can be targeted for deletion.
We can delete the entire coding sequence of a gene using CRISPR/Cas9. You can then study the phenotype of the true null allele with no concern that any protein function remains. Our transgenic designers and process ensure that each knockout we make is exactly the right strain to answer the research question.
Partial Gene Deletion
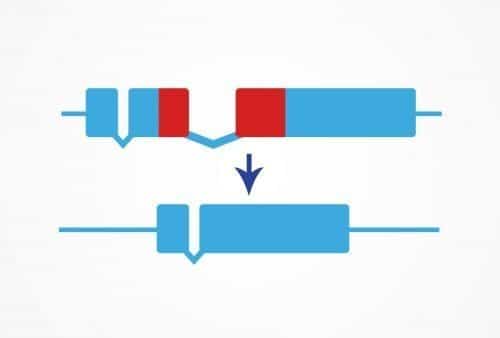
The Precise Deletion service uses CRISPR/Cas9 genome editing to remove a small defined region of DNA. This can be useful for deleting protein domains, DNA regulatory regions, or any other sequence of your choice.
Deletions can also be performed in coding regions for functional analysis of protein domains or to isolate isoform function. Transcription factor binding sites or introns can be deleted to reveal information about gene regulation.
Conditional Gene Deletion
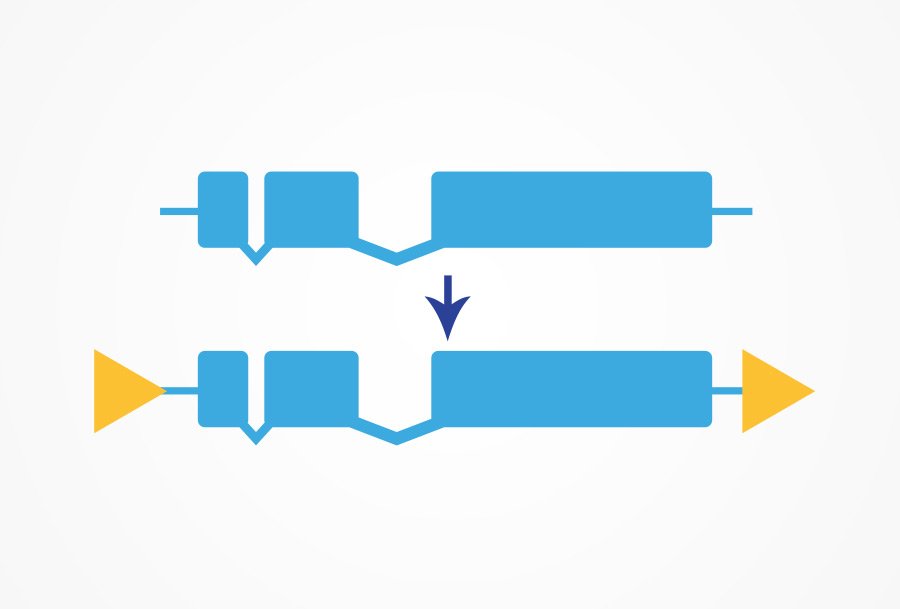
Floxed alleles can be created for a tissue- or temporal- specific deletion of your gene of interest. This is a powerful tool for studying embryonic lethal genes or for understanding how your gene of interest functions in different tissues. CRISPR/Cas9 is used to insert loxP sites flanking the region to be deleted.
This line can then be crossed with a Cre-line expressing the Cre recombinase under a specific promoter or injected with a plasmid containing Cre recombinase. The Cre recombinase promotes recombination of the two loxP sites and the region between the sites is removed from the genome.
Conditional alleles
- Used to make spatiotemporal gene deletions
- Flank gene of interest with LoxP sites
- Inject a plasmid or cross into strain that expresses Cre recombinase where/when/how you want loss-of-function to occur
We are able to build custom Cre recombinase-expressing lines to compliment your new floxed allele.
CRISPR Knock-out Case Study
A client wanted a knockout (KO) of an embryonic lethal gene. We could not make this line using our standard methods. Instead, we inserted two loxP sites. One in the first intron of the gene and the second in the 3’utr. After we confirmed this line by PCR and sequencing, we injected this line with a ubiquitously expressing Cre Recombinase plasmid.
We found while the uninjected animals could reproduce, the Cre injected animals did not (Figure A). We tested recombination by PCR and found that only the Cre injected progeny showed recombination of the loxP sites (Figure B).
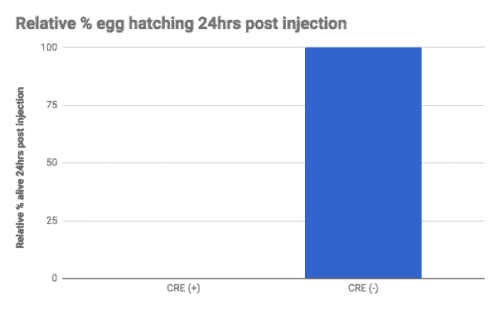
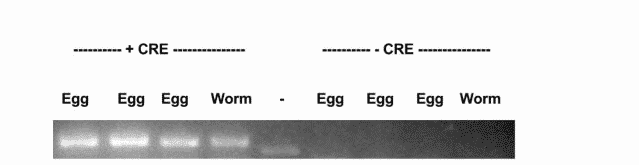
This line could be used to study KO of this gene in adulthood or in specific tissues, something that was not previously possible due to the embryonic lethality caused by the KO of this gene.
C. elegans Crispr Knockout Service Pricing
InVivo Biosystems offers a comprehensive pricing structure for its C. elegans Knockout Service, providing affordable options for researchers interested in gene editing. With various package options available, researchers can choose the pricing plan that best suits their individual needs and budget.
Service Package
Price
Est. Delivery Time
Full Build
$3,465 and up
8 weeks and up
Candidate Lines
$2,435 and up
6 weeks and up
Custom Injection Mix
$1,740 and up
4 weeks and up
FAQs About CRISPR Knockout
CRISPR Knockout uses a system of enzymes and RNA to cut and disable a specific gene in a cell or organism.
CRISPR Knockout offers high efficiency, specificity, versatility, speed, and cost-effectiveness compared to traditional gene editing techniques.
CRISPR Knockout has numerous applications in disease modeling, drug discovery, and gene therapy development, as well as basic genetics research.
Researchers use CRISPR Knockout to study the function of specific genes, create disease models, develop new drugs, and advance our understanding of gene function and disease mechanisms.
Various companies and research organizations offer CRISPR Knockout services such as InVivo Biosystems, providing researchers and scientists with access to this cutting-edge technology.
The development of CRISPR Knockout technology has paved the way for more advanced gene editing techniques, including CRISPR activation, base editing, and prime editing, which will continue to advance genetic research and biotechnology.
Publications
Ready to get started?
Ready to connect with us to learn more about working with our company or our technology?
Submit your inquiry below & we will get back to you soon.

